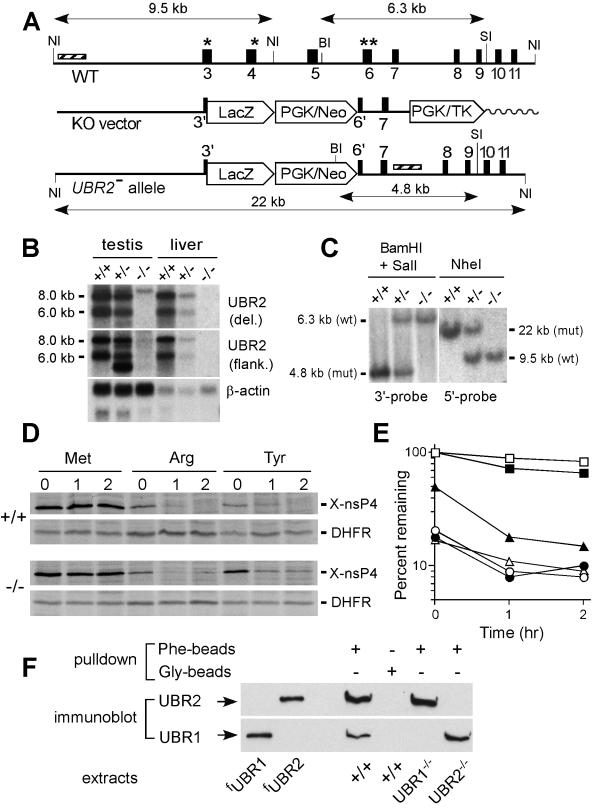
FIG. 2.
Construction of UBR2−/− mouse strains. (A) Targeting vector. A map of the ∼30-kb 5′-proximal region of the ∼96-kb m-UBR2 gene, the targeting vector, and the deletion disruption UBR2−/− allele. Exons are designated by vertical rectangles. Solid and wavy horizontal lines designate, respectively, the mouse and plasmid DNAs. The directions of gene transcription are indicated as well. Exons 3 and 4, marked by single asterisks, and exon 6, marked by a double asterisk, contain conserved residues that are essential for integrity of the substrate-binding sites of sc-UBR1. NI, NheI; BI, BamHI; SI, SalI. Southern hybridization probes are indicated by striped rectangles. PGK, phosphatidylglycerol kinase; TK,thymidine kinase. (B) Northern analysis of m-UBR2 expression with total RNA from the testes and livers of adult +/+, UBR+/−, and UBR2−/− mice. The probes used were either the 350-bp UBR2 cDNA fragment (nt 383 to 731) that was deleted in the UBR2−/− allele (data not shown) or the 2.3-kb UBR2 cDNA fragment (nt 863 to 3227) adjacent to the deleted region (upper panel) or the human β-actin cDNA fragment (lower panel). The wild-type 8.0- and 6.0-kb UBR2 mRNAs are indicated on the left. (C) Southern analysis of BamHI/SalI-cut (3′ probe) and NheI-cut (5′ probe) mouse tail DNA. The 760-bp BamHI/SalI 3′ probe detected 6.3- and 4.8-kb UBR2 fragments for the wild-type (wt) and mutant (mut) UBR2 alleles, respectively. The 1.2-kb NheI 5′ probe detected 9.5-kb (wild type) and 22-kb (mutant) fragments. (D) Pulse-chase analysis with +/+ and UBR2−/− EF cell lines. EFs were transfected with plasmids expressing fDHFRh-UbR48-X-nsP4f, which yielded the fDHFRh-UbR48 reference protein (designated DHFR), and an X-nsP4f (X-nsP4-Flag) test protein (X = Met, Arg, or Tyr) (designated X-nsP4). Cells were labeled for 10 min with [35S]methionine and chased for 1 and 2 h. (E) Quantitation of the patterns in shown in panel D with PhosphorImager. For each time point, the ratio of 35S in X-nsP4f to 35S in the Met-DHFR-UbR48 reference protein at the same time point was plotted as the percentage of that ratio relative to that for Met-nsP4f (which bore a stabilizing N-terminal residue) at time zero (the beginning of chase). Open and closed symbols designate the results with +/+ and UBR2−/− EFs, respectively. Squares, Met-nsP4f; circles, Arg-nsP4f; triangles, Tyr-nsP4f. (F) Extracts from +/+, UBR1−/−, and UBR2−/− EFs were assayed (see Results) by using peptide pull-down assays and immunoblotting with anti-UBR1 and anti-UBR2 antibodies. fUBR1 and fUBR2 designate S. cerevisiae extracts expressing Flag-tagged m-UBR1 and m-UBR2.