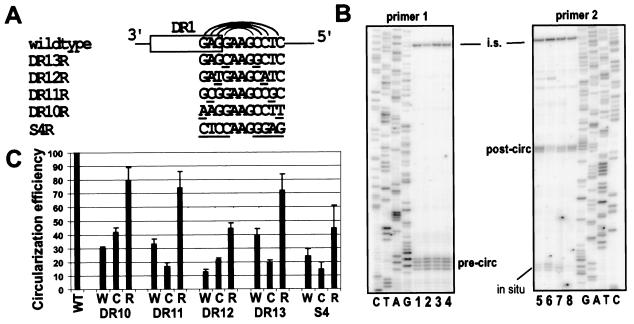
FIG. 2.
Primer extension analyses of variants indicated that the presence of the hairpin increases circularization efficiency. (A) Substitutions were introduced into the stem nucleotides of the hairpin to initially reduce (W and C variants) and subsequently restore (R variants) to mutant sequence the potential for Watson-Crick base pairing. The R variant for each set of mutations is shown, with the substituted nucleotides in both sides of the stem underscored. The W and C variants contained the same substitutions as the R variants introduced into either the 3′ or 5′ half of the stem, respectively. Sequence is indicated in the minus-strand orientation. (B) Primer extension analyses on DNA replication intermediates isolated from LMH cells using primers 1 and 2 with samples from the DR13 series. Samples included internal standard DNA. Primer 1 detected plus-strand DNA that initiated from DR2 and elongated, at least, to the 5′ end of the minus-strand DNA (pre-circ). Primer 2 detected the fraction of plus-strand DNA detected by primer 1 that successfully circularized and elongated (post-circ). Primer 1 also detected plus strands initiating from DR1 (in situ). Detection of the internal standard (i.s.) is shown near the top of both gel images. The sequencing ladders were generated using the same primers as in the respective primer extension reactions. Lanes 1 and 8, wild-type reference; lanes 2 and 7, DR13W; lanes 3 and 6, DR13C; lanes 4 and 5, DR13R. (C) The circularization efficiency for each variant was calculated as described in the legend for Fig. 1B. Samples were normalized to a wild-type reference (set to 100) and are presented as means with error bars to indicate the standard deviations. Each virus was analyzed multiple times (n = 2 to 7) from independent transfections.