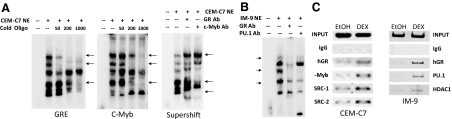
Figure 6.
Analysis of the Protein-Binding Sites in the −4559/−4525 GRU
EMSAs (panels A and B) were performed as described in Ref. 11 and ChIP assays were done as described in Ref. 12. A, EMSAs were performed using nuclear extracts of CEM-C7 cells that were treated with 1 μm DEX for 24 h. The extracts were incubated with a 32P-labeled −4559/−4525 GRU oligonucleotide, either alone or in the presence of increasing molar fold excesses of an unlabeled consensus GRE (left panel) or c-Myb consensus sequence (middle panel). Arrows indicate protein/GRU complexes that were competed out by the cold competitor oligonucleotides. In the right panel, the nuclear extract was incubated with the labeled GRU in the presence of no antibody or antibodies to the GR or c-Myb. Complexes that were altered by the antibodies are indicated by the arrows. B, EMSA supershift analysis was performed with nuclear extracts from IM-9 cells that were treated with 1 μm DEX for 24 h. Arrows indicate protein-DNA complexes that were altered by antibodies to the GR and PU.1. C, ChIP analysis. CEM-C7 (left panel) and IM-9 (right panel) cells were treated for 24 h with 1 μm DEX. ChIP analysis was performed with antibodies to the indicated proteins. Ab, Antibody; NE, nuclear extract.