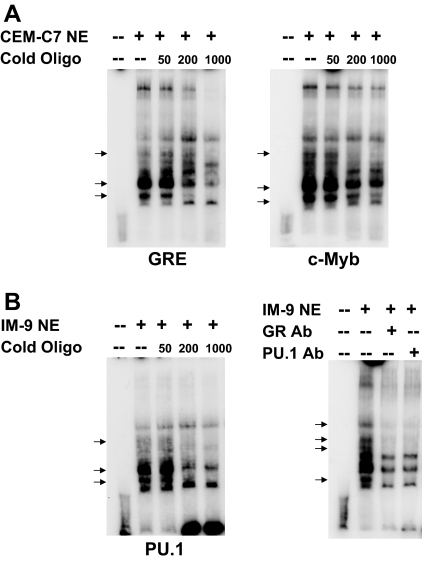
Figure 9.
EMSA of the −2956/−2916 GRU
EMSAs were performed as described in Ref. 11. A, EMSAs were performed using nuclear extracts of CEM-C7 cells that were treated with 1 μm DEX for 24 h. The extracts were incubated with a 32P-labeled −2956/−2916 GRU oligonucleotide, either alone or in the presence of increasing molar fold excesses of an unlabeled consensus GRE (left panel) or c-Myb consensus sequence (right panel). Arrows indicate protein/GRU complexes that were competed out by the cold competitor oligonucleotides. B, EMSAs were performed using nuclear extracts of IM-9 cells that were treated with 1 μm DEX for 24 h. The extracts were incubated with a 32P-labeled −2956/−2916 GRU oligonucleotide, either alone or in the presence of increasing molar fold excesses of an unlabeled PU.1 consensus sequence (left panel). Arrows indicate protein/GRU complexes that were competed out by the cold competitor oligonucleotides. EMSA supershift analysis (right panel) was performed with nuclear extracts from IM-9 cells that were treated with 1 μm DEX for 24 h. Arrows indicate protein-DNA complexes that were altered by antibodies to the GR and PU.1. Ab, Antibody; NE, nuclear extract.