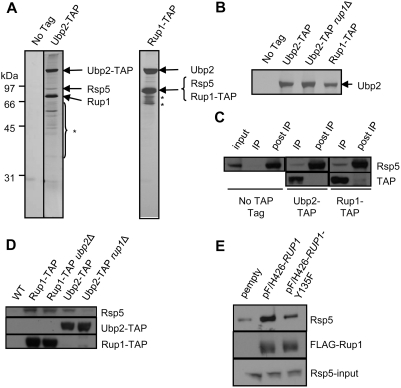
Figure 1. Ubp2, Rsp5, and Rup1 interact physically.
(A) Silver-stained SDS polyacrylamide gel showing affinity purified Ubp2-TAP and Rup1-TAP protein complexes. Arrows indicate polypeptides identified by gel band excision followed by MALDI-ToF mass spectrometry. The asterisk indicates degradation products of Ubp2 and Rup1. (B) Ubp2 recovery from Ubp2-TAP, Ubp2-TAP rup1Δ, and Rup1-TAP strains. Arrow indicates gel band identified as Ubp2 by MALDI-ToF mass spectrometry. (C) Co-immunoprecipitation of a fraction of cellular Rsp5 with Ubp2 and Rup1. Cell extracts were depleted of endogenously TAP-tagged Ubp2 or Rup1 with IgG (IP lanes), and the remaining soluble Rsp5 (post-IP lanes) and bait proteins probed by Western blot with anti-Rsp5 antibodies which also recognized the protein A portion of the TAP tag. The ‘input’ lane shows the Rsp5 level in the extract prior to IP. (D) Rup1 tethers Ubp2 to Rsp5. IgG-based immunoprecipitation of Rup1-TAP and Ubp2-TAP from WT, ubp2Δ or rup1Δ strains. The co-purification of Rsp5 with the baits was determined by Western blotting using anti-Rsp5 antibodies. (E) Plasmids expressing FLAG(F/H)-tagged wildtype Rup1 or the Y135F point mutant were transformed into cells lacking endogenous Rup1. After immunoprecipitation with anti-FLAG antibodies, Rup1 and Rsp5 were detected by Western blotting. The bottom panel show the expression levels of Rsp5 in whole cell extracts by Western blotting against Rsp5.