Abstract
Context: Interindividual variations in glucocorticoid sensitivity have been associated with manifestations of cortisol excess or deficiency and may be partly explained by polymorphisms in the human glucocorticoid receptor (hGR) gene. We studied a 43-yr-old female, who presented with manifestations consistent with tissue-selective glucocorticoid hypersensitivity. We detected a novel, single, heterozygous nucleotide (G → C) substitution at position 1201 (exon 2) of the hGR gene, which resulted in aspartic acid to histidine substitution at amino acid position 401 in the amino-terminal domain of the hGRα. We investigated the molecular mechanisms of action of the natural mutant receptor hGRαD401H.
Methods-Results: Compared with the wild-type hGRα, the mutant receptor hGRαD401H demonstrated a 2.4-fold increase in its ability to transactivate the glucocorticoid-inducible mouse mammary tumor virus promoter in response to dexamethasone but had similar affinity for the ligand (dissociation constant = 6.2 ± 0.6 vs. 6.1 ± 0.6 nm) and time to nuclear translocation (14.75 ± 0.25 vs. 14.25 ± 1.13 min). The mutant receptor hGRαD401H did not exert a dominant positive or negative effect upon the wild-type receptor, it preserved its ability to bind to glucocorticoid response elements, and displayed a normal interaction with the glucocorticoid receptor-interacting protein 1 coactivator.
Conclusions: The mutant receptor hGRαD401H enhances the transcriptional activity of glucocorticoid-responsive genes. The presence of the D401H mutation may predispose subjects to obesity, hypertension, and other manifestations of the metabolic syndrome.
A novel mutation of the human glucocorticoid receptor (hGR) gene enhances its gene expression and may predispose subjects to obesity, hypertension, and metabolic syndrome-related atherosclerotic cardiovascular disease.
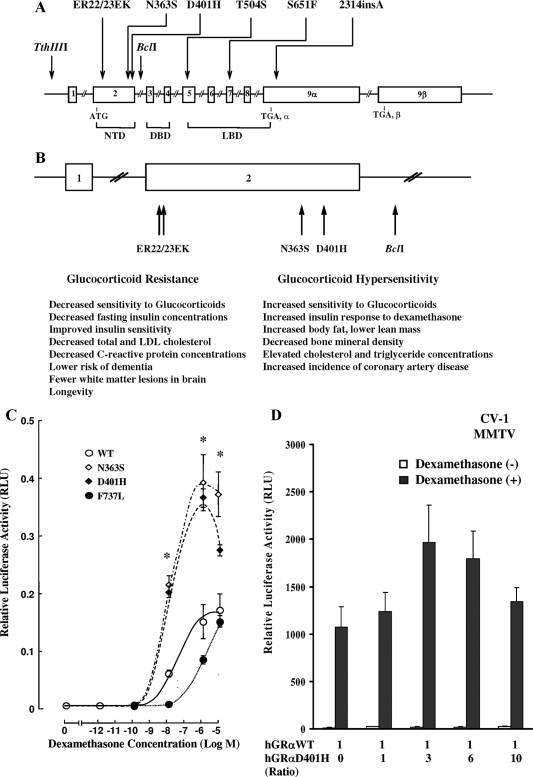
Glucocorticoids regulate a broad spectrum of physiological functions essential for life (1,2). At the cellular level, their actions are mediated by the glucocorticoid receptor (GR), a ligand-dependent transcription factor (3). Tissue sensitivity to glucocorticoids varies considerably among individuals within a healthy population and may be partly influenced by polymorphisms in the human GR (hGR) gene (nuclear receptor subfamily 3, group C, member 1; gene identification: 2908) (4). At least three polymorphisms of the hGR gene, the N363S, the BclI restriction fragment length polymorphism, and the ER22/23EK, have been reported to result in alterations in glucocorticoid sensitivity, body composition, and metabolic parameters (Fig. 1, A and B). We present a novel hGR mutation associated with manifestations of glucocorticoid hypersensitivity.
Figure 1.
A, Schematic representation of the location of the reported polymorphisms in the hGR gene. B, Schematic representation of the hGR gene polymorphisms located in the amino-terminal domain of the receptor and a summary of their clinical associations (4,13,14,15,16,17,18,19). C, Transcriptional activity of the wild-type (WT) hGRα and mutant receptors hGRαD401H, hGRαN363S, and hGRαF737L. Compared with the wild-type receptor, the mutant receptor hGRαD401H demonstrated a 2.4-fold increase in its ability to transactivate the MMTV promoter in response to dexamethasone. Y bars represent sem. The asterisks indicate significant differences in the transcriptional activity between the wild-type hGRα and mutant receptors hGRαD401H and hGRαN363S (P < 0.05). D, Absence of a dominant positive or negative effect of the mutant receptor hGRαD401H upon the wild-type hGRα. Cotransfection with a constant amount of hGRα and progressively increasing concentrations of hGRαD401H resulted in an additive increase of hGRα-mediated transactivation of the MMTV promoter. Bars represent mean ± sem of at least five independent experiments. Solid bars indicate treatment with dexamethasone (10−6 m), whereas open bars indicate no treatment with dexamethasone. No statistically significant differences were observed in the mean relative luciferase activity (RLU) values between different hGRα to hGRαD401H ratios. DBD, DNA-binding domain; LBD, ligand-binding domain; LDL, low-density lipoprotein; NTD, amino-terminal domain.
A 43-yr-old Colombian female presented with a 10-yr history of clinical manifestations consistent with tissue-selective glucocorticoid hypersensitivity, including visceral obesity, dyslipidemia, type 2 diabetes, and hypertension. Physical examination revealed normal body mass index (24.2 kg/m2), elevated waist to hip ratio (0.96; normal range < 0.85), and no manifestations of Cushing’s syndrome. Endocrinological evaluation at the National Institutes of Health revealed elevated 0800-h serum cortisol (33.0 μg/dl) and plasma ACTH (127.0 pg/ml) concentrations, but normal urinary free cortisol excretion (52 μg/24 h; 37 μg/24 h). An ovine CRH (oCRH) test showed robust cortisol and ACTH responses to oCRH stimulation (peak cortisol: 49.1 μg/dl; peak ACTH: 480 pg/ml). Five years before her presentation, the patient had been diagnosed with epilepsy and been treated with carbamazepine. Carbamazepine has been associated with increased serum cortisol concentrations and 24 h urinary free cortisol excretion, as well as robust ACTH and cortisol responses to oCRH stimulation in healthy volunteers, an effect that is likely to reflect relative insufficiency of glucocorticoid negative feedback at the pituitary level (5). Furthermore, carbamazepine is a hepatic enzyme inducer known to result in false-positive dexamethasone suppression tests, thereby leading to misdiagnosis of Cushing’s syndrome (6). Following written informed consent, genomic DNA was obtained, and the hGR gene was amplified and sequenced. We identified a novel, single, heterozygous nucleotide (G → C) substitution at position 1201 in exon 2 of the gene, which resulted in aspartic acid to histidine substitution at amino acid position 401 in the amino-terminal domain of the hGRα (nuclear receptor subfamily 3, group C, member 1 isoform α; protein identification: NP000167). No other mutations or polymorphisms were identified. We investigated the molecular mechanisms of action of the natural mutant receptor hGRαD401H.
Materials and Methods
Plasmids
The plasmids used included pRShGRα, pRShGRαD401H, pRShGRαN363S, pRShGRαF737L, pF25GFP-hGRα, pF25GFP-hGRαD401H, pBK/CMV-hGRα, pBK/CMV-hGRαD401H, pGEX4T3-GRIP1 (1–1462), pGEX4T3-GRIP1 (596–774), pGEX4T3-GRIP1 (740–1217), pRSV-erbA−1, pMMTV-luc, and pSV40-β-gal (7–11).
Transactivation assays
CV-1 cells were cotransfected with pRShGRα pRShGRαN363S, pRShGRαF737L, or pRShGRαD401H (0.05 μg/well), pMMTV-luc (0.5 μg/well), and pSV40-β-gal (0.1 μg/well). In further experiments, cells were cotransfected with pMMTV-luc, pSV40-β-gal, a constant amount of pRShGRα, and progressively increasing concentrations of pRShGRαD401H. A control plasmid was added in appropriate quantities to maintain a constant amount of DNA in each well. Cells were exposed to dexamethasone for 24 h, and luciferase and β-galactosidase activities were determined in the cell lysates (7,8,9,10,11). Luciferase activity was divided by β-galactosidase activity to account for transfection efficiency.
Western blot analyses
CV-1 and COS-7 cells were transfected with pRShGRα or pRShGRαD401H (15 μg/flask). Western blot analyses were performed as previously described (7,8,9,10,11).
Dexamethasone-binding assays
Dexamethasone-binding assays were performed on peripheral blood mononuclear cells obtained from the patient and a control subject. Ligand-binding assays were also performed on COS-7 cells transfected with pRShGRα or pRShGRαD401H (1.5 μg/well) (7,8,9,10,11).
Nuclear translocation studies
HeLa cells were transfected with pF25GFP-hGRα or pF25GFP-hGRαD401H (2 μg/dish). In further experiments, cells were transfected with equal amounts of pF25GFP-hGRα and pRShGRαD401H (1.5 μg/dish). Nuclear translocation studies were performed as previously described (7,8,9,10,11).
Chromatin immunoprecipitation (ChIP) assays
HCT-116 cells, in which the mouse mammary tumor virus (MMTV) promoter was stably integrated within chromatin, were transiently transfected with pRShGRα or pRShGRαD401H (10 μg/dish). ChIP assays were performed as previously described (8,9,10,11).
Glutathione-S-transferase (GST) pull-down assays
In vitro transcription/translation reactions were used to produce 35S-labeled hGRα and hGRαD401H in rabbit reticulocyte lysate by using pBK/CMV-hGRα and pBK/CMV-hGRαD401H, respectively, as templates. The in vitro interaction between hGRα-related plasmids and GST-fused GR-interacting protein 1 (GRIP1) proteins was tested as previously described (7,8,9,10,11).
Results
The mutant receptor hGRαD401H displays increased transcriptional activity compared with the wild-type hGRα
Compared with the wild-type hGRα, the mutant receptor hGRαD401H demonstrated a 2.4-fold increase in its ability to transactivate the glucocorticoid-inducible MMTV promoter in response to dexamethasone (10−12 to 10−5 m) (Fig. 1C). This effect of hGRαD401H was similar to that of hGRαN363S, a polymorphism known to be associated with glucocorticoid hypersensitivity, but opposite to the effect of hGRαF737L, a natural mutant receptor associated with glucocorticoid resistance (7,8,9,10,11). The concentration of dexamethasone required to achieve 50% of transactivation was 10−8 m for the mutant receptor and 14.5 × 10−8 m for the wild-type receptor.
The mutant receptor hGRαD401H does not exert a dominant positive or negative effect upon the wild-type hGRα
Cotransfection with a constant amount of hGRα and progressively increasing concentrations of hGRαD401H indicated that the mutant receptor hGRαD401H had an additive rather than a dominant positive or negative effect upon the wild-type receptor (Fig. 1D).
The mutant receptor hGRαD401H demonstrates normal affinity for the ligand
The apparent dissociation constant of hGRαD401H was similar to that of hGRα in COS-7 cells transfected with the respective plasmids [6.2 ± 0.6 vs. 6.1 ± 0.6 nm; P = not significant (NS)]. Furthermore, there was no difference in the affinity of hGRα for the ligand between the patient and the control subject in peripheral blood mononuclear cells (dissociation constant, 8.95 ± 2.3 vs. 8.39 ± 3.11 nm; P = NS). No difference in the number of dexamethasone-binding sites was noted between the wild-type and mutant receptor in both assays.
Western blot analyses demonstrated no differences in the expression of hGRα and hGRαD401H proteins in CV-1 or COS-7 cells, indicating that the aforementioned results did not reflect differences at the protein expression level.
The mutant receptor hGRαD401H demonstrates normal subcellular localization and nuclear translocation
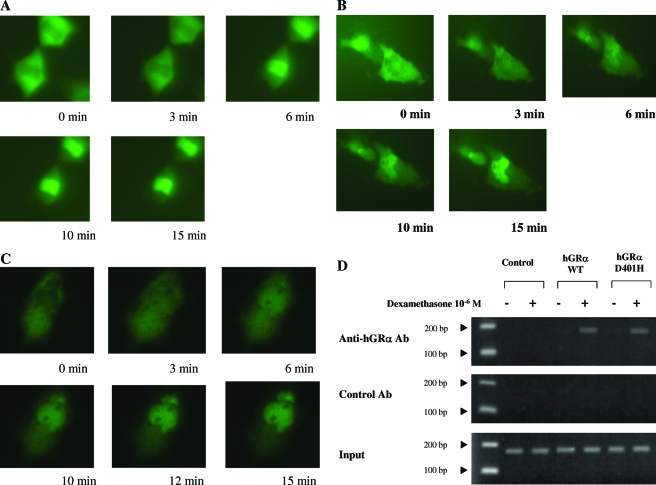
In the absence of dexamethasone, both the wild-type and mutant receptors were primarily localized in the cytoplasm of cells. No significant differences were observed between hGRα and hGRαD401H in the time required for nuclear translocation after exposure to dexamethasone (10−6 m) (14.75 ± 0.25 vs. 14.25 ± 1.13 min; P = NS) (Fig. 2, A and B). Coexpression of hGRα and hGRαD401H at a 1:1 ratio had no apparent effect on the nuclear translocation of the wild-type hGRα (Fig. 2C).
Figure 2.
Nuclear translocation of pF25GFP-hGRα (A) pF25GFP-hGRαD401H (B), and pF25GFP-hGRα in the presence of pRShGRαD401H (C) before and after exposure to dexamethasone. HeLa cells transiently expressing pF25GFP-hGRα or pF25GFP-hGRαD401H were exposed to the same concentration of dexamethasone (10−6 m). Images of the same cells were obtained at the indicated time points. There were no statistically significant differences between the wild-type (WT) hGRα and mutant receptor hGRαD401H in the time required for nuclear translocation after exposure to the ligand. D, ChIP assays performed on HCT-116 cells, in which the MMTV promoter was stably integrated into the chromatin. Both hGRα and hGRαD401H coprecipitated with MMTV glucocorticoid response elements in a ligand-dependent fashion, indicating that the mutant receptor hGRαD401H preserves its ability to bind to DNA. Ab, Antibody.
The mutant receptor hGRαD401H preserves its ability to bind to DNA in vivo
In ChIP assays, both hGRα and hGRαD401H coprecipitated with MMTV glucocorticoid response elements similarly in a ligand-dependent fashion, suggesting that hGRαD401H preserves its ability to bind to DNA (Fig. 2D).
The mutant receptor hGRαD401H interacts with the GRIP1 coactivator in vitro through both its activation function (AF)-1 and AF-2 domains
GRIP1 contains two sites that bind to steroid receptors: one site interacts with the AF-2 of hGRα in a ligand-dependent fashion, whereas the other site interacts with the AF-1 of hGRα in a ligand-independent fashion (12). In GST pull-down assays, like the wild-type receptor, hGRαD401H interacted with the GRIP1 coactivator through both its AF-1 and AF-2 domains (Fig. 2D).
Discussion
We identified a novel, heterozygous point mutation in exon 2 of the hGR gene and investigated the molecular mechanisms through which the mutant receptor affects glucocorticoid signal transduction. We demonstrated that the D401H mutation enhanced the hGRα-mediated transactivation of glucocorticoid-responsive genes but did not affect the affinity of the receptor for the ligand, its cytoplasmic to nucleus translocation, its ability to bind to DNA, or its interaction with the GRIP1 coactivator. Furthermore, the mutant receptor hGRαD401H had an additive rather than a dominant positive or negative effect upon the wild-type receptor. Our findings suggest that, even at a heterozygotic state, the D401H mutation enhances the transcriptional activity of hGRα and may predispose carriers to an adverse metabolic profile, which, along with other important determinants, such as environmental, dietary, and socioeconomic factors, may lead to visceral obesity, hypertension, dyslipidemia, and metabolic syndrome-related atherosclerotic cardiovascular disease.
Interindividual variations in tissue sensitivity to glucocorticoids have been described within the normal population and have been partly attributed to polymorphisms in the hGR gene. Several polymorphisms of the hGR gene have been reported (4). The N363S polymorphism is associated with higher sensitivity to glucocorticoids in vivo, increased insulin response to exogenous dexamethasone administration, higher body mass index and waist to hip ratio, elevated cholesterol and triglyceride concentrations, lower bone mineral density in trabecular bone, and higher incidence of coronary artery disease independent of weight (4,13,14,15).
A frequent BclI restriction fragment length polymorphism is also associated with increased sensitivity to glucocorticoids, hypertension, visceral adiposity, and increased insulin concentrations in obese women (4,16).
A third polymorphism, the ER22/23EK, is associated with relative glucocorticoid resistance, lower fasting insulin concentrations and improved insulin sensitivity, lower total and low-density lipoprotein cholesterol concentrations, and lower C-reactive protein concentrations (4,17,18). In line with this favorable metabolic profile, the ER22/23EK polymorphism is significantly higher in the oldest half of the population and is associated with increased survival. Furthermore, at older age, carriers of this polymorphism have lower risk of dementia and fewer white matter lesions in the brain compared with noncarriers (4).
We believe that the D401H mutation in our patient is producing a mixed glucocorticoid resistance and hypersensitivity phenotype with tissue selectivity. Therefore, the presence of resistance at the regulatory centers of the hypothalamic-pituitary-adrenal (HPA) axis is associated with hypercortisolism, whereas the presence of hypersensitivity at the vasculature, central adipose tissue, and liver is associated with hypertension, central adiposity, and dyslipidemia. Interestingly, the N363S polymorphism was also associated with a paradoxically hyperresponsive HPA axis at the Trier social stress test, suggesting that this point mutation caused glucocorticoid resistance at the level of feedback regulation of the HPA axis (19).
Like most polymorphic variants, the D401H mutation is located at the amino-terminal domain of the receptor, in close proximity to the major transactivation domain AF-1. Given that modification of AF-1 by phosphorylation may enhance or repress the transcriptional activity of hGRα in a gene- and tissue-specific manner, it is possible that the D401H mutation causes a tissue-specific effect on AF-1 activity (20). This hGR gene mutation has not been previously described, and its prevalence in Colombia or other countries has not been determined. Therefore, it is essential that it should be included in population studies aiming to detect hGR variants associated with glucocorticoid hypersensitivity.
In clinical practice, glucocorticoids are used widely to treat a number of pathological conditions, as well as for replacement therapy purposes. The effects of glucocorticoid treatment may vary considerably between patients and may be partly attributed to polymorphisms in the hGR gene. Therefore, when the presence of these hGR gene variants is known in a patient, the dose of glucocorticoids should be adjusted accordingly to ensure optimal therapy and minimal adverse effects (4).
We conclude that the D401H mutation enhances the hGRα-mediated transactivation of glucocorticoid-responsive genes in a tissue-specific manner, and may predispose subjects to obesity, hypertension, and metabolic syndrome-related atherosclerotic cardiovascular disease.
Footnotes
This work was funded by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, the European Union-European Social Fund, and the Greek Ministry of Development-General Secretariat of Research and Technology, Athens, Greece.
Disclosure Statement: The authors have nothing to disclose.
First Published Online September 30, 2008
Abbreviations: AF, Activation function; ChIP, chromatin immunoprecipitation; GR, glucocorticoid receptor; GRIP1, glucocorticoid receptor-interacting protein 1; GST, glutathione-S-transferase; hGR, human glucocorticoid receptor; HPA, hypothalamic-pituitary-adrenal; MMTV, mouse mammary tumor virus; NS, not significant.
References
- Chrousos GP, Charmandari E, Kino T 2004 Glucocorticoid action networks—an introduction to systems biology. J Clin Endocrinol Metab 89:563–564 [DOI] [PubMed] [Google Scholar]
- Charmandari E, Tsigos C, Chrousos G 2005 Endocrinology of the stress response. Annu Rev Physiol 67:259–284 [DOI] [PubMed] [Google Scholar]
- Zhou J, Cidlowski JA 2005 The human glucocorticoid receptor: one gene, multiple proteins and diverse responses. Steroids 70:407–417 [DOI] [PubMed] [Google Scholar]
- van Rossum EF, Lamberts SW 2004 Polymorphisms in the glucocorticoid receptor gene and their associations with metabolic parameters and body composition. Recent Prog Horm Res 59:333–357 [DOI] [PubMed] [Google Scholar]
- Perini GI, Devinsky O, Hauser P, Gallucci WT, Theodore WH, Chrousos GP, Gold PW, Kling MA 1992 Effects of carbamazepine on pituitary-adrenal function in healthy volunteers. J Clin Endocrinol Metab 74:406–412 [DOI] [PubMed] [Google Scholar]
- Ma RC, Chan WB, So WY, Tong PC, Chan JC, Chow CC 2005 Carbamazepine and false positive dexamethasone suppression tests for Cushing’s syndrome. BMJ 330:299–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmandari E, Kino T, Souvatzoglou E, Vottero A, Bhattacharyya N, Chrousos GP 2004 Natural glucocorticoid receptor mutants causing generalized glucocorticoid resistance: molecular genotype, genetic transmission, and clinical phenotype. J Clin Endocrinol Metab 89:1939–1949 [DOI] [PubMed] [Google Scholar]
- Charmandari E, Raji A, Kino T, Ichijo T, Tiulpakov A, Zachman K, Chrousos GP 2005 A novel point mutation in the ligand-binding domain (LBD) of the human glucocorticoid receptor (hGR) causing generalized glucocorticoid resistance: the importance of the C terminus of hGR LBD in conferring transactivational activity. J Clin Endocrinol Metab 90:3696–3705 [DOI] [PubMed] [Google Scholar]
- Charmandari E, Chrousos GP, Ichijo T, Bhattacharyya N, Vottero A, Souvatzoglou E, Kino T 2005 The human glucocorticoid receptor (hGR) β isoform suppresses the transcriptional activity of hGRα by interfering with formation of active coactivator complexes. Mol Endocrinol 19:52–64 [DOI] [PubMed] [Google Scholar]
- Charmandari E, Kino T, Ichijo T, Zachman K, Alatsatianos A, Chrousos GP 2006 Functional characterization of the natural human glucocorticoid receptor (hGR) mutants hGRαR477H and hGRαG679S associated with generalized glucocorticoid resistance. J Clin Endocrinol Metab 91:1535–1543 [DOI] [PubMed] [Google Scholar]
- Charmandari E, Kino T, Ichijo T, Jubiz W, Mejia L, Zachman K, Chrousos GP 2007 A novel point mutation in helix 11 of the ligand-binding domain of the human glucocorticoid receptor gene causing generalized glucocorticoid resistance. J Clin Endocrinol Metab 92:3986–3990 [DOI] [PubMed] [Google Scholar]
- Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR 1998 Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol 12:302–313 [DOI] [PubMed] [Google Scholar]
- Huizenga NA, Koper JW, De Lange P, Pols HA, Stolk RP, Burger H, Grobbee DE, Brinkmann AO, De Jong FH, Lamberts SW 1998 A polymorphism in the glucocorticoid receptor gene may be associated with an increased sensitivity to glucocorticoids in vivo. J Clin Endocrinol Metab 83:144–151 [DOI] [PubMed] [Google Scholar]
- Lin RC, Wang XL, Morris BJ 2003 Association of coronary artery disease with glucocorticoid receptor N363S variant. Hypertension 41:404–407 [DOI] [PubMed] [Google Scholar]
- Rosmond R, Chagnon YC, Holm G, Chagnon M, Pérusse L, Lindell K, Carlsson B, Bouchard C, Björntorp P 2000 A glucocorticoid receptor gene marker is associated with abdominal obesity, leptin, and dysregulation of the hypothalamic-pituitary-adrenal axis. Obes Res 8:211–218 [DOI] [PubMed] [Google Scholar]
- Weaver JU, Hitman GA, Kopelman PG 1992 An association between a Bc1I restriction fragment length polymorphism of the glucocorticoid receptor locus and hyperinsulinaemia in obese women. J Mol Endocrinol 9:295–300 [DOI] [PubMed] [Google Scholar]
- van Rossum EF, Koper JW, Huizenga NA, Uitterlinden AG, Janssen JA, Brinkmann AO, Grobbee DE, de Jong FH, van Duyn CM, Pols HA, Lamberts SW 2002 A polymorphism in the glucocorticoid receptor gene, which decreases sensitivity to glucocorticoids in vivo, is associated with low insulin and cholesterol levels. Diabetes 51:3128–3134 [DOI] [PubMed] [Google Scholar]
- van Rossum EF, Feelders RA, van den Beld AW, Uitterlinden AG, Janssen JA, Ester W, Brinkmann AO, Grobbee DE, de Jong FH, Pols HA, Koper JW, Lamberts SW 2004 Association of the ER22/23EK polymorphism in the glucocorticoid receptor gene with survival and C-reactive protein levels in elderly men. Am J Med 117:158–162 [DOI] [PubMed] [Google Scholar]
- Wüst S, Van Rossum EF, Federenko IS, Koper JW, Kumsta R, Hellhammer DH 2004 Common polymorphisms in the glucocorticoid receptor gene are associated with adrenocortical responses to psychosocial stress. J Clin Endocrinol Metab 89:565–573 [DOI] [PubMed] [Google Scholar]
- Kino T, Ichijo T, Amin ND, Kesavapany S, Wang Y, Kim N, Rao S, Player A, Zheng YL, Garabedian MJ, Kawasaki E, Pant HC, Chrousos GP 2007 Cyclin-dependent kinase 5 differentially regulates the transcriptional activity of the glucocorticoid receptor through phosphorylation: clinical implications for the nervous system response to glucocorticoids and stress. Mol Endocrinol 21:1552–1568 [DOI] [PubMed] [Google Scholar]