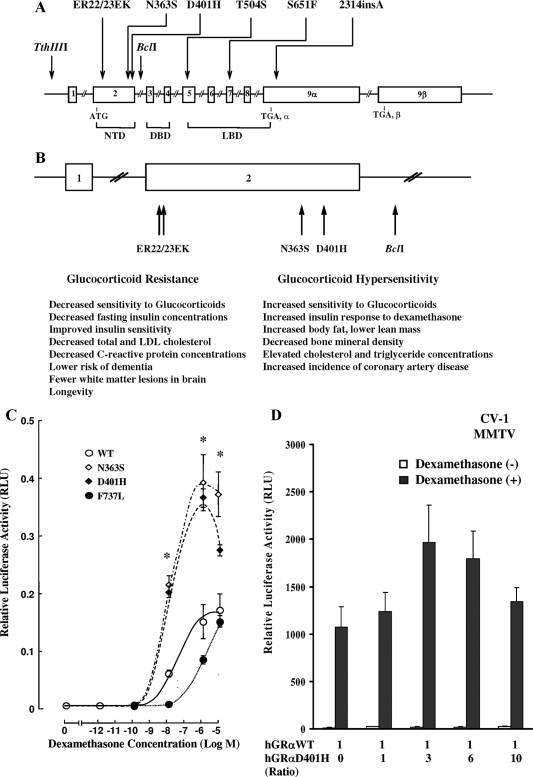
Figure 1.
A, Schematic representation of the location of the reported polymorphisms in the hGR gene. B, Schematic representation of the hGR gene polymorphisms located in the amino-terminal domain of the receptor and a summary of their clinical associations (4,13,14,15,16,17,18,19). C, Transcriptional activity of the wild-type (WT) hGRα and mutant receptors hGRαD401H, hGRαN363S, and hGRαF737L. Compared with the wild-type receptor, the mutant receptor hGRαD401H demonstrated a 2.4-fold increase in its ability to transactivate the MMTV promoter in response to dexamethasone. Y bars represent sem. The asterisks indicate significant differences in the transcriptional activity between the wild-type hGRα and mutant receptors hGRαD401H and hGRαN363S (P < 0.05). D, Absence of a dominant positive or negative effect of the mutant receptor hGRαD401H upon the wild-type hGRα. Cotransfection with a constant amount of hGRα and progressively increasing concentrations of hGRαD401H resulted in an additive increase of hGRα-mediated transactivation of the MMTV promoter. Bars represent mean ± sem of at least five independent experiments. Solid bars indicate treatment with dexamethasone (10−6 m), whereas open bars indicate no treatment with dexamethasone. No statistically significant differences were observed in the mean relative luciferase activity (RLU) values between different hGRα to hGRαD401H ratios. DBD, DNA-binding domain; LBD, ligand-binding domain; LDL, low-density lipoprotein; NTD, amino-terminal domain.