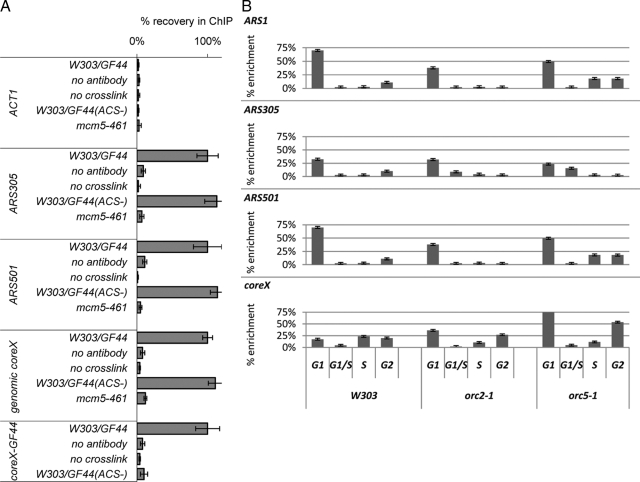
Figure 6.
(A) Mcm5p association with telomeres depends on ACS. ChIP with anti-Mcm5p antibodies (Santa Cruz Biotechnology) was performed in the W303/GF44, W303/GF44(ACS−), and mcm5-461 strains. Each experiment was performed with parallel controls where cross-linking or primary antibody incubation was omitted. The abundance of different DNAs (shown on the left) was quantified by PCR after confirming linear PCR range for the input samples. The primers for the core X element in GF44 and GF44(ACS−) anneal to the plasmid backbone of the integrating constructs and do not amplify genomic core X. core X signals were also confirmed by slot-blot. The signals in the eluates were subtracted by the signals in the final washes and then divided by the signals in the input fractions to produce “net signals.” Finally, all net signals were normalized so that the corresponding values in GF44 are 100% and then compared with the values in W303/GF44(ACS−) and mcm5-461. The figure is representative of two independent ChIP experiments. (B) Wild-type (W303), orc2-1 and orc5-1 cells (shown at the bottom top) were released from mitotic arrest and samples were collected after 10, 35, 60, and 75 min (early G1, late G1/S, S, and G2 phases of the cell cycle, respectively) as shown at the bottom. ChIP with anti-Mcm5p antibodies was conducted as described above. The signals in the eluates were subtracted by the signals in the final washes and then divided by the signals in the input fractions and plotted as % enrichment relative to input. The analyzed genomic loci are indicated in the top left corner of each graph. The figure is representative of two independent ChIP experiments.