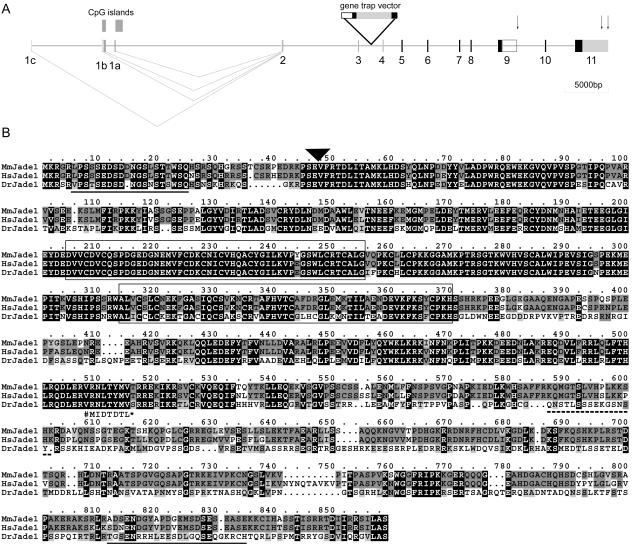
FIG. 5.
Jade1 gene structure and multiple-protein alignment of mouse, human, and zebra fish Jade1. (A) Mouse Jade1 is encoded by 10 exons (black boxes 2 to 11). The gene trap insertion occurred between exons 3 and 4. Noncoding exons are shown in numbered grey boxes. Potential polyadenylation signals are indicated by arrows. The box outlined at the end of exon 9 indicates an alternative 3′ untranslated region sequence that would give rise to a ∼3.5-kb transcript and the short Jade1 isoform (Jade1S), equivalent to the published human Jade1 protein (69). EST and cDNA alignments indicate that at least three alternative 5′ noncoding exons (1a to 1c) are used. A promoter scan identified two putative promoter regions in proximity to exons 1a and 1b that coincide with CpG islands. (B) Multiple alignment of mouse Jade1 (Mm) with the putative Jade1 orthologues in human (Hs) and zebra fish (Dr) reveals extensive homology. Residues identical in all three homologues are filled black while those identical only in two of these and conserved amino acid substitutions are filled dark and light grey, respectively. The PHD finger domains are boxed. A potential bipartite NLS is indicated by a dashed underline. Although this NLS is not conserved in the zebra fish, two possible overlapping bipartite NLS are present at positions 635 to 651 and 648 to 664 of DrJade1. Two putative PEST sequences (PESTFIND score > 10) found in both mouse and human Jade1 are underlined in solid black. The position of the gene trap insertion is indicated by the black triangle. The published human Jade1 protein sequence (GenPept accession no. AAM95612) is shorter than the sequence shown here; AAM95612 is identical up to the position marked # below the aligned sequences (corresponding to the end of exon 9) but is truncated at a stop codon (*) after 7 variant amino acids encoded by intron 9.