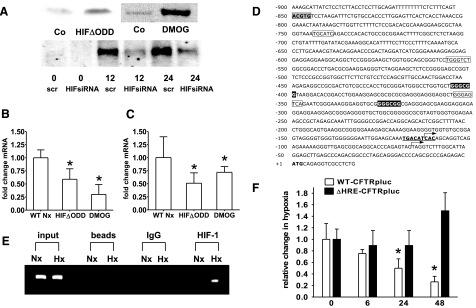
Figure 3.
CFTR repression is mediated by HIF-1. A) Western blot analysis of nuclear Caco-2 protein samples. Constitutive HIF-1 activation was achieved by using cells expressing HIFΔODD or following hydroxylase inhibition (DMOG; 1 mM for 24 h). B, C) CFTR mRNA levels in Caco-2 (B) and T84 cells (C) under conditions of normoxic HIF-1 activation. Results are means ± se from 3 separate experiments; *P ≤ 0.05 vs. control, which integrates results from respective controls for each HIF-1 intervention. D) Sequence of the CFTR promoter; numbering is relative to the initiation codon (boldface). Putative hypoxia responsive element (gray box), AP-1 sites (boxed), GC-box (inverted type), and a binding site for CREB (bold, underlined) are highlighted. Arrows indicate the tissue-specific transcription start sites identified for the small intestine and intestinal epithelial cell lines (27, 43). E) ChIP demonstrates HIF-1 binding to genomic DNA corresponding to the CFTR 5′-UTR in normoxic (Nx) and hypoxic (Hx) Caco-2 cells (24 h). Reaction controls included PCR performed on genomic DNA (input), on samples precipitated by protein A sepharose beads (beads) or by beads with IgG. F) CFTR promoter activity was examined with luciferase constructs driven by the CFTR promoter. WT (WT-CFTRpLuc) was compared to a promoter construct bearing a mutation in the HIF-1 binding site (ΔHRE-CFTRpLuc). Promoter activity was evaluated following indicated periods of hypoxia in Caco-2 cells. Results are calculated from the ratio of luciferase-activity to activity of cotransfected control plasmid (renilla), presented as means ± se from 3 experiments; n = 3/condition; *P ≤ 0.05 vs. normoxia.