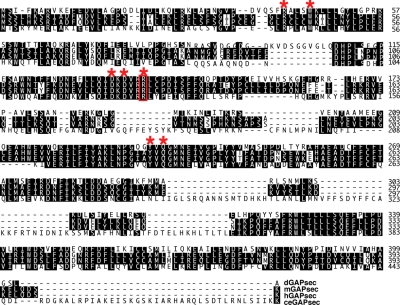
Figure 1.
Sequence comparison of Drosophila dGAPsec protein with orthologs of mouse, human and C. elegans. Conserved amino acids (black background) and GAP designating residues (red asterisks) are highlighted. Red box, essential arginine residue used to generate dominant-negative GAPsec mutations (see text). Top to bottom: dGAPsec, mGAPsec (Tbc1d13; NM_146252), human hGAPsec (Tbc1d13; NM_018201) and C. elegans ceGAPsec (F45E6.3; NM_077694).