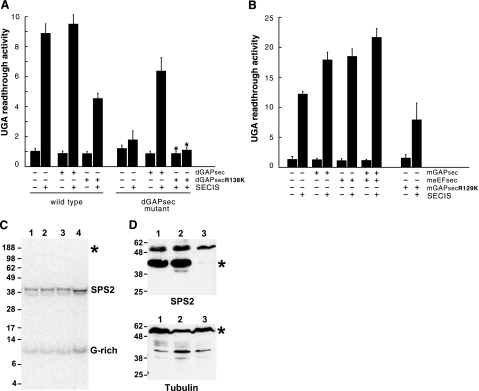
Figure 3.
Loss of GAPsec activity affects SECIS-dependent UGA read-through in Drosophila and mouse but not selenocysteine incorporation. A) Lack of SECIS-dependent read-through in dGAPsec mutant flies. Activities of the lacZ/UGA/luciferase reporter gene (RG) with or without the SECIS element (SECIS) in wild-type and dGAPsec mutants after dGAPsec and dGAPsecR138K overexpression, respectively (details in the text). “UGA read-through activity” refers to fold increase of luciferase activity over the basis level in cells containing the reporter gene without SECIS sequences (15, 17, 21). Note that SECIS-dependent UGA read-through is impaired in dGAPsec mutants and in wild-type flies expressing dGAPsecR138K. Note also that the lack of read-through in dGAPsec mutants can be rescued by dGAPsec but not by dGAPsecR138K expression. B) UGA read-through in transfected mouse NIH 3T3 cells. Bars represent means ± sd from 6 independent experiments; for rescue experiments, mean values and limits of 2 independent experiments are shown (∗). meEFsec (B) served as the response control of the assay system (details in text). +, presence of indicated activity; −, absence of indicated activity. C) Selenoprotein synthesis of wild-type and dGAPsec mutant flies after 75Se labeling. Numbers on the left of the SDS-PAGE gel refer to sizes (kDa) of marker proteins. Lane 1: wild-type flies expressing the RG containing the SECIS-element; lane 2, wild-type flies that express only β-galactosidase (RG lacks the SECIS element); lane 3, dGAPsec mutant flies without RG; and lane 4, dGAPsec mutant flies that express only β-galactosidase (RG contains the SECIS element). Note selenoprotein synthesis (SPS2 and G-rich) in all lanes and the absence of 75Se label in the β-galactosidase/luciferase fusion protein (position indicated by ∗), showing that the fusion protein (15, 17, 21) lacks 75Se incorporation (details in text). D) Western blot containing proteins of wild-type flies (lane 1) and dGAPsec (lane 2) and deEFsec mutants (lane 3) using anti-SPS2 antibodies (top panel) and anti-tubulin antiserum (bottom panel). Numbers on the left refer to sizes (kDa) of marker proteins. ∗, positions of SPS2 and tubulin (loading control).