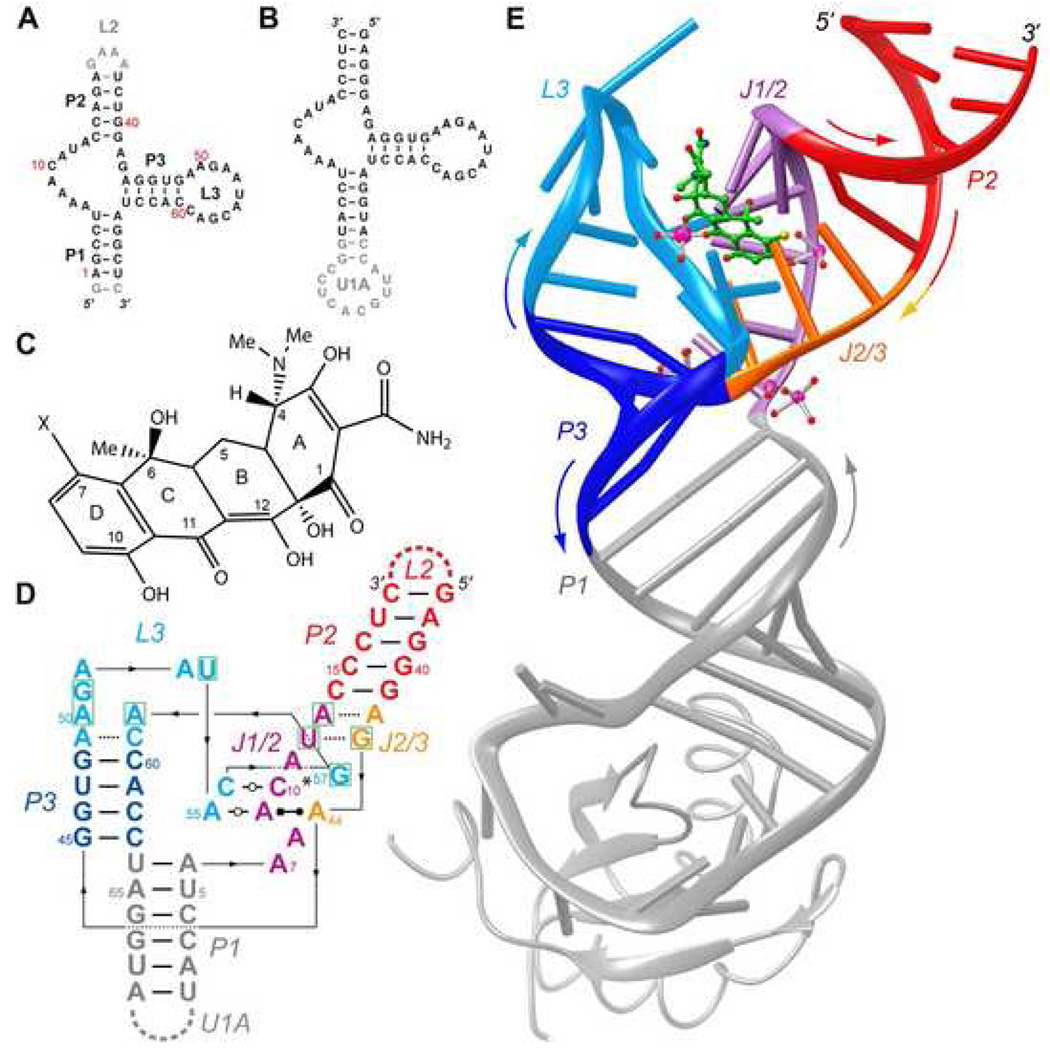
Figure 1. Overall structure of the tetracycline aptamer.
A, Sequence of a wild-type connectivity tetracycline aptamer. Residue numbers correspond to those of the constructs of Hanson et al. (2005) and Müller et al. (2006). Gray nucleotides are functionally dispensable. B, Sequence of the circularly-permuted crystallization construct. Note deletion of loop L2, and introduction of an U1A protein binding site capping paired region P1. C, Chemical structure of tetracycline, numbered conventionally. X is hydrogen or chlorine, for tetracycline and 7-chlorotetracycline, respectively. D, Schematic secondary structure of the tetracycline-bound conformation of the aptamer. Nucleotides that form the antibiotic binding site are boxed in green. Thin lines with embedded arrowheads depict connectivity. Watson-Crick and single-hydrogen bond pairs are indicated by thin and dashed lines, respectively. Non-canonical base pair symbols are those of Leontis and Westhof (2001). Asterisk indicates that the base of G57 makes single hydrogen bonds with the bases of both, C10 and A11. E, Cartoon representation of the tetracycline aptamer (color-coded as in panel D) in complex with 7-chlorotetracycline (green stick figure) and U1A. Selected well-ordered magnesium ions and their solvation waters (magenta and red spheres, respectively) are shown. Colored arrows denote chain direction.