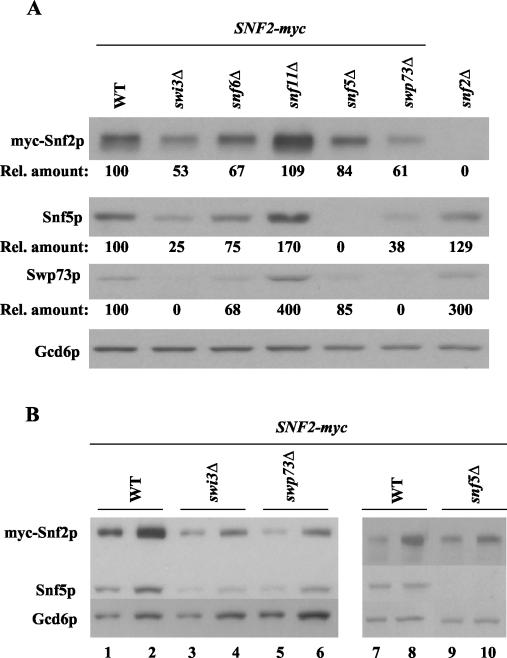
FIG. 6.
Western blot analysis of SWI/SNF subunits in swi/snf mutants. (A) The GCN4 strains containing SNF2-myc and either WT SWI/SNF subunits or the indicated swi/snf deletions were grown in SC-ILV, and total proteins were extracted as described in Materials and Methods. Aliqouts with equal amounts of total proteins were separated by SDS-PAGE by using 10% gels, transferred to a nitrocellulose membrane, and probed with antibodies against myc, Snf5p, or Swp73p, as indicated on the left of the blot. Probing with Gcd6p antibodies provided a loading control. The Western blotting signals were quantified by video densitometry by using NIH Image software, normalized for the Gcd6p signals, expressed relative to the normalized value measured in WT cells, and listed below the corresponding blots for myc-Snf2p, Snf5p, and Swp73p (Rel. amount). (B) The swp73Δ and swi3Δ strains exhibit similar reductions in SWI/SNF subunit levels. The analysis described for panel A was repeated for the WT, swi3Δ, swp73Δ, and snf5Δ strains harboring SNF2-myc, loading two different amounts of extract in adjacent lanes; thus, the sizes of samples in lanes 1, 3, 5, 7, and 9 were 50% of those loaded in lanes 2, 4, 6, 8, and 10.