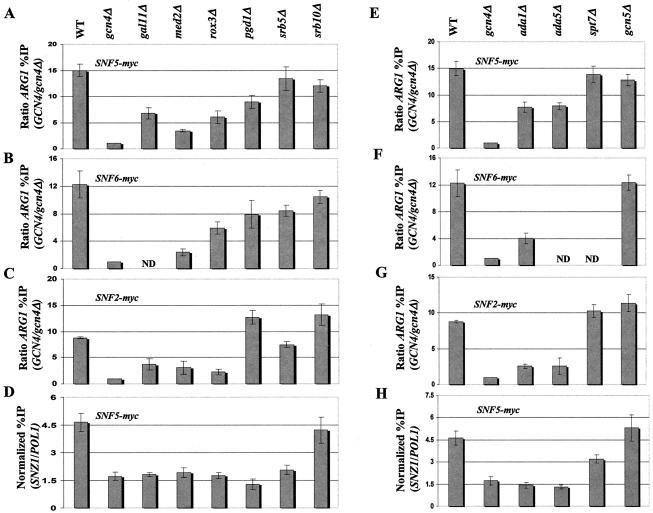
FIG. 9.
Subunits of mediator and SAGA are required for optimal recruitment of SWI/SNF by Gcn4p. (A through C and E through G) We generated a panel of strains harboring the chromosomal myc-tagged SWI/SNF alleles designated in the upper left of each histogram from GCN4 strains containing all WT coactivator subunits (WT) or the indicated deletion alleles (shown at the top) that remove subunits of mediator (A through C) or SAGA (E through G) and from the gcn4Δ strain containing all WT coactivator subunits (gcn4Δ) (see Table 1 for a list of strains). High-copy-number plasmid pHQ1239 was introduced into the GCN4 strains, and empty vector YCp50 was introduced into the gcn4Δ strains. The resulting transformants were subjected to ChIP analysis to measure binding of the myc-tagged SWI/SNF subunit to ARG1, as described in the legend to Fig. 1. (D and H) The strains described for panels A and E that harbor the SNF5-myc allele were subjected to ChIP analysis to measure binding of myc-Snf5p to SNZ1. ND, not determined.