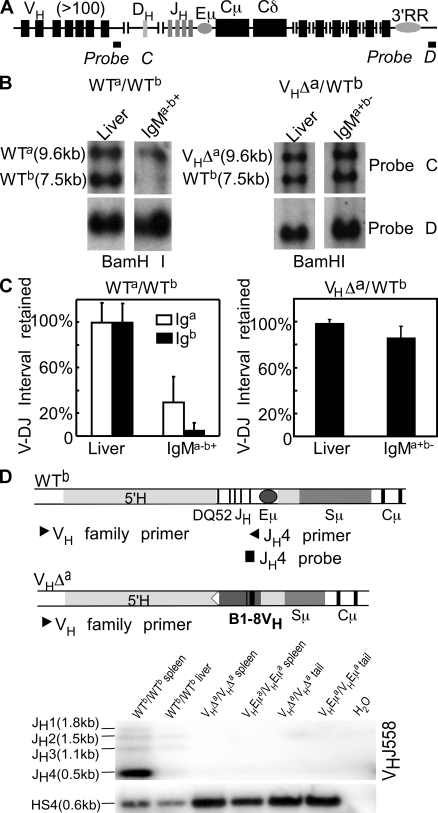
Figure 4.
V-DJ rearrangements on the WT Ighb allele of VHΔa/WTb mice. (A) Igh locus. Probe C detects a sequence deleted upon V-DJ joining. Probe D detects a sequence retained on both alleles in all cells. (B) Assay for V-DJ rearrangement on the Igha and Ighb alleles in WTa/WTb and VHΔa/WTb mice. Genomic DNA extracted from liver and from sorted IgMa−b+ spleen cells (WTa/WTb; left) and IgMa+b− spleen cells (VHΔa/WTb; right). DNAs were digested with BamHI and hybridized with probes C (top) and D (bottom). (C) Quantitative analysis (ImageQuant) of blot shown in B. A repeat experiment was done using liver and spleen cells from VHΔa/WTb animals. Band intensities in liver were set to 100%. Normalization was to probe D for WTa/WTb mice; normalization was to the Igha fragment in VHΔa/WTb mice. Error bars show SD. (D) Igh locus maps indicating PCR primers and probes for detecting V-DJ rearrangements on Ighb allele. 5′ primers specific for individual VH gene families (VH family primer) were used in combination with a 3′ primer (JH4 primer), which was present on the Ighb allele but missing on both the VHEμa and VHΔa alleles. The representative blot demonstrates allele-specific amplification. H2O, no DNA template control.