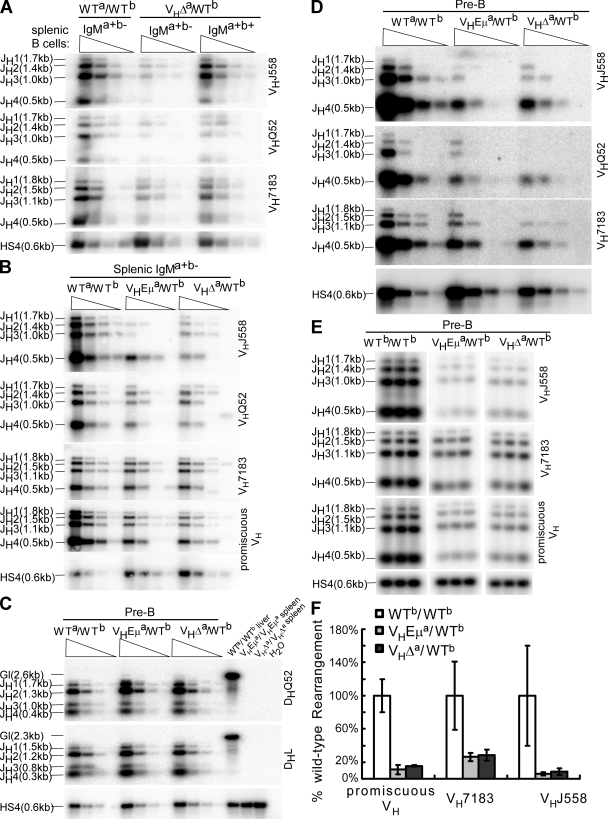
Figure 5.
V-DJ rearrangements on the WT Ighb allele of VHEμa/WTb and VHΔa/WTb mice. (A) Splenic B cells from WTa/WTb and VHΔa/WTb, sorted on the basis of phenotype (IgMa+b− or IgMa+b+). PCRs were performed on fivefold serial dilutions of DNA templates (four lanes/cell type). PCR strategy detects V-DJ rearrangements on only the Ighb allele in VHΔa/WTb mice (both alleles in WTa/WTb mice). VDJ rearrangements involving each JH gene segment are indicated (JH1, JH2, etc.). VH family primers are indicated to the right of blots (VHJ558, etc.). PCR of HS4 (a 3′ RR element) was normalized for DNA input. (B) Like A, but a primer designed to anneal to all VH families (promiscuous VH 46) was included. (C) D-J rearrangements on the Ighb allele of pre-B cells (B220+CD43−IgM−) sorted from six mice of each genotype. The DHL primer anneals to most DH genes (47); DQ52 anneals only to DHQ52. Allele-specific 3′ primer (JH4) was used as in A. Liver DNA from WTa/WTb mice and spleen cell DNA from homozygous VHEμa and VHΔa mice were included as controls. H2O, no template control. (D) Pre-B cells were isolated as in C and PCR reactions using the VH gene family primers were performed as described in A. (E) Pre-B cells were isolated as in C. Sample dilutions determined to be in the linear range (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20081202/DC1) were prepared in triplicate for PCR. Representative data is shown. (F) Quantitative analysis of VDJ rearrangement blots. For each PCR reaction with a given VH primer, signals for JH1, JH2, JH3, and JH4 rearrangements were quantified and summed by ImageQuant. Values obtained for pre-B cells from VHEμa/WTb and VHΔa/WTb mice were normalized to the corresponding values for pre-B cells of WTb/WTb mice. Data were obtained from two pools of two mice each for WTb/WTb and three pools involving a total of seven mice for each of the VHEμa/WTb and VHΔa/WTb mouse lines. Error bars show SD.