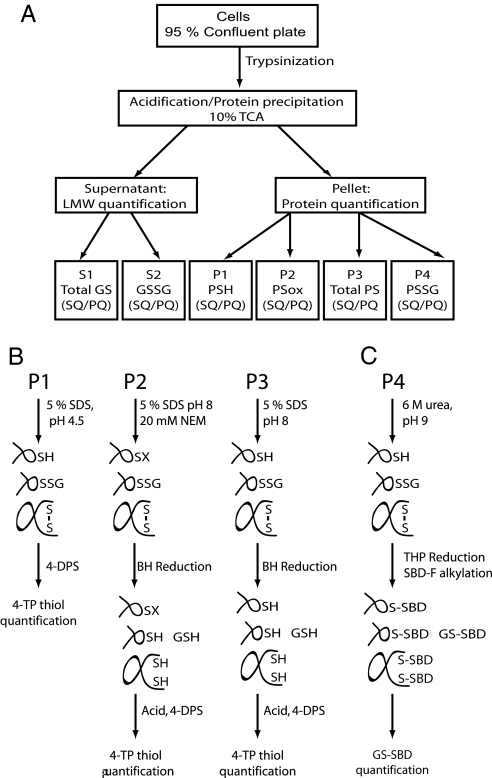
Fig. 1.
Experimental approach for quantifying the cellular thiol/disulfide environment. (A) Flow chart defining individual fractions quantified. To avoid contribution of protein disulfides from serum in the growth medium, cells were trypsinized and washed in PBS before addition of TCA to 10%. After centrifugation, the supernatant contains low-molecular-weight thiol species, and proteins are pelleted. The supernatant is divided in two for the quantification of total (S1) and oxidized (S2) glutathione. The TCA pellet is divided into 4 samples, used for quantification of the following redox species: PSH (P1); PSox = PSSP + PSSG (P2), Total PS (P3), and PSSG (P4). In addition to redox species, the total protein content in each pellet is quantified and used as a common denominator for the individual samples. SQ, sulfhydryl quantification; PQ, protein quantification. (B) Quantification of protein thiols and disulfides. Pellets were solubilized by sonication in 5% SDS buffers appropriate for either thiol or disulfide quantification. PSH was quantified from P1 by use of 4-DPS, essentially as described in ref. 12. PSox was quantified from P2 by first alkylating PSH with NEM followed by disulfide reduction with BH. Before thiol quantification with 4-DPS, BH was destroyed by addition of acid. As a control, Total PS was experimentally measured by solubilizing P3 in SDS without NEM and subjecting the sample to BH reduction and 4-DPS quantification, as described above. (C) Quantification of PSSG. Glutathione bound to protein (P4) was modified by reaction with the fluorogenic thiol label SBD-F under reducing conditions. The SBD-GS derivative was subsequently detected by HPLC as described in SI Text. This method quantitatively detected GSSG exogenously added to P4 samples, confirming the reliability of the method.