Abstract
Telomeres are nucleoprotein structures that cap the ends of chromosomes and thereby protect their stability and integrity. In the presence of telomerase, the enzyme that synthesizes telomeric repeats, telomere length is controlled primarily by Rap1p, the budding yeast telomeric DNA binding protein which, through its C-terminal domain, nucleates a protein complex that limits telomere lengthening. In the absence of telomerase, telomeres shorten with every cell division, and eventually, cells enter replicative senescence. We have set out to identify the telomeric property that determines the replicative capacity of telomerase-deficient budding yeast. We show that in cells deficient for both telomerase and homologous recombination, replicative capacity is dependent on telomere length but not on the binding of Rap1p to the telomeric repeats. Strikingly, inhibition of Rap1p binding or truncation of the C-terminal tail of Rap1p in Kluyveromyces lactis and deletion of the Rap1p-recruited complex in Saccharomyces cerevisiae lead to a dramatic increase in replicative capacity. The study of the role of telomere binding proteins and telomere length on replicative capacity in yeast may have significant implications for our understanding of cellular senescence in higher organisms.
Telomeres are the specialized DNA-protein structures at the ends of eukaryotic chromosomes essential for chromosomal stability and integrity (3, 4, 52). In most eukaryotes, telomeric DNA is synthesized by telomerase which copies a short template sequence within its own RNA moiety (17, 54). Telomere binding proteins of various organisms share a primary role; to negatively regulate telomere length in the presence of telomerase (11, 30, 59). It has been suggested that a telomere length-sensing system counts the number of telomeric repeats or proteins bound to them and translates this information to the machineries which add or deplete telomeric repeats (43, 55).
Understanding how telomere length is monitored has significant implications especially for the processes of aging and cancer; most higher eukaryotic cells that can divide cannot do so indefinitely. Thus, normal cells are said to have a finite life span (23). The process that limits the proliferative potential of cells has been termed cellular or replicative senescence (34). How do cells sense the number of divisions through which they have gone? In somatic cells, telomerase activity is repressed. In addition, conventional DNA polymerases are unable to fully replicate the ends of chromosomes due to their primer dependency. As a result, telomeres of all the somatic cells and tissues tested shorten with replicative age (1, 22). At present telomere shortening is perhaps the most viable explanation of a cell division counting mechanism (2, 12, 21). A causal relationship between telomere shortening and in vitro cellular senescence was established, as the reactivation of telomerase in cultured cells was directly demonstrated to result in an extended life span leading to their apparent immortalization (6). The limited replicative potential of somatic cells, dictated by telomere shortening, can act as a tumor suppression mechanism. It has been shown that replenishing telomeres by an active telomerase is one of the few essential steps which a normal human fibroblast must take on its way to becoming cancerous (19).
Progressive shortening of telomeres in the absence of telomerase results in the generation of chromosome end-to-end fusions in late generation mTR−/− mice and in cell cycle arrest and p53-mediated apoptosis (5, 9, 25, 28, 32). The complete loss of the telomeric repeats of a few critically short telomeres was shown to limit cellular survival in telomerase-deficient mice (26). In primary cells, the failure to protect critically shortened telomeres leads to replicative senescence (29). In mammalian cells, it was suggested that it is not telomere length per se but the interaction of TRF2 with telomeric DNA which determines the minimal telomere length required for continuation of cell doublings (29). It was recently suggested that the reduction in the amount of telomeric single-stranded DNA might be the actual cause for replicative senescence (58). Many aspects of telomere structure and length control are shared between all eukaryotic organisms studied.
Yeast cells defective in the telomerase pathway, as the Saccharomyces cerevisiae mutants Δtlc1 and Δest1-4 or the Kluyveromyces lactis telomerase mutant Δter1 show a phenotype very similar to that seen in mammalian cells: progressive telomere shortening accompanied by gradual loss of cell viability through a senescence-like mechanism (33, 37, 45, 56).
In the absence of telomerase, two alternative recombination pathways collaborate to maintain the telomeres of S. cerevisiae. In the absence of RAD52, both pathways are inactivated and no cells survive (31). In K. lactis, in the absence of telomerase, the production of postsenescence survivors was also shown to be strongly dependent on the RAD52 gene (44).
In the presence of telomerase, the binding of the double-stranded DNA binding protein Rap1p and its associated interacting factors to the telomeric repeats is central for many telomere-associated functions in yeast. Among these are telomere length regulation, telomeric silencing, and meiosis (8, 10, 30, 38-41). The use of Rap1p C-terminal deletion mutants and artificial tethering of the C terminus of Rap1p to the telomere as well as the use of mutants, which reduce Rap1p binding to the telomere, has shed much light on the mode of telomere length control in the presence of telomerase (47). Currently, the role of Rap1p in determining the replicative capacity of yeast cells deficient for telomerase and homologous recombination is unknown. Theoretically, it could be either the number of telomere-bound Rap1p molecules or alternatively, the length of the telomeric DNA that is critical for determination of a functional telomere, which allows continuation of cell division of yeast cells in the absence of telomere maintenance activities.
To examine these possibilities, we have constructed an experimental system to monitor the fate of yeast cells deficient for telomerase. Using this system, we could show that the binding of Rap1p or the presence of its C-terminal domain in K. lactis and the RAP-RIF complex in S. cerevisiae are dispensable for the replicative capacity of yeast.
MATERIALS AND METHODS
Southern blots.
Genomic DNA was prepared from saturated cultures by using a modified version of the Zymolase method and hybridized as described previously (41). A K. lactis-specific telomeric probe was prepared by phosphorylation with T4 polynucleotide kinase of a telomeric G-strand oligonucleotide (5′-GGTATGTGGTGTACGGATTTGATTA-3′). An S. cerevisiae-specific telomeric probe was prepared by phosphorylation with T4 polynucleotide kinase of a telomeric G-strand oligonucleotide (5′-TGGGTGTGGGTGTGGGTGTGGGTG-3′). Hybridizations (1 h) and washes (total 15 min) were performed at 45°C.
Plasmid and strain construction. (i) K. lactis plasmids.
pRG3, a multi-copy-number plasmid carrying TER1, was constructed by ligation of a 4-kb BamHI-XbaI TER1 fragment to the BamHI-XbaI site of the pCXJ3 vector. ter1-AA, ter1-Bsi, and ter1-Acc were subcloned into pCXJ3 in a similar manner to create the ter1 multi-copy-number vectors pSM7, pSM8, and pSM11, respectively. pJR32 and pJR24, carrying TER1 on low-copy-number URA3 and HIS3 CEN-ARS plasmids, respectively, were provided by Elizabeth Blackburn.
(ii) K. lactis strains.
The Δter1 strain was constructed by transformation of a K7B520 strain with the ter1-Δ7 integrative plasmid (provided by Mike McEachern) and screening for the ter1-Δ7 pop-out by PCR. The Δter1 rap1-ΔC strain was constructed by the pop-in pop-out method with an integrative plasmid carrying the rap1-ΔC allele and screening for the rap1-ΔC pop-out by PCR.
The Δrad52 Δter1 rap1ΔC and Δrad52 Δter1 RAP1 K. lactis strains were created by mating a K7B520-based Δrad52 strain (46) with the Δter1 rap1-ΔC strain. The resulting diploid was sporulated. The Δrad52 Δter1 rap1ΔC and Δrad52 Δter1 RAP1 spore products were screened by PCR. To create RAP1 or rap1-ΔC strains carrying the low-copy-number TER1 allele, the Δrad52 Δter1 RAP1 or Δrad52 Δter1 rap1-ΔC strain was transformed by pJR24. To create RAP1 strains carrying the ter1 alleles, TER1, ter1-AA, ter1-Bsi, and ter1-Acc, the Δrad52 Δter1 RAP1 strain was transformed by the high-copy-number plasmids pRG3, pSM7, pSM8, and pSM11, respectively, to create the ySM132, ySM133, ySM134, and ySM135 strains respectively. To create Δrad52 rap1-ΔC strains carrying the various ter1 alleles TER1, ter1-AA, ter1-Bsi, and ter1-Acc, the Δrad52 Δter1 rap1-ΔC strain was transformed by pRG3, pSM7, pSM8, and pSM11, respectively, to create the ySM136, ySM137, ySM138, and ySM139 strains, respectively.
S. cerevisiae plasmid and strain construction. (i) S. cerevisiae plasmids.
pSM2, an S. cerevisiae CEN-ARS plasmid carrying RAD52 and TLC1 was constructed by subcloning a 3.3-kb SalI fragment of RAD52 in the XhoI site of pRS316 and a 2.1-kb BamHI-XhoI fragment of TLC1 in the BamHI-SalI sites of this vector. pSM4, an S. cerevisiae CEN-ARS plasmid carrying RAD52 and tlc1-C476G was created by ligating a 1.3-kb tlc1-C476G fragment from pRS306-tlc1-C476G (provided by Elizabeth Blackburn) to the pRS316 vector cut by XhoI.
(ii) S. cerevisiae strains.
ySM2, a Δtlc1 Δrad52 strain carrying a CEN-ARS TLC1 plasmid, and ySM11, a Δtlc1 Δrad52 strain carrying a CEN-ARS tlc1-C476G plasmid, were created by transforming pSM2 or pSM4, respectively, to a tlc1::TRP1 rad52::LEU2 strain based on W303a (provided by Elizabeth Blackburn). A Δrif1 strain was created by mating a Δrif1 strain in which RIF1 has been disrupted with the KanMX cassette provided by Research Genetics (in a 4741 background) with a Δtlc1 Δrad52 strain carrying a CEN-ARS TLC1 plasmid that was backcrossed four times with 4742.
Determination of replicative capacity.
Cells were plated on 5-fluoroorotic acid (5-FOA) to select for loss of the TER1 or TLC1 plasmid. When HIS3 plasmids were used, colonies were plated on yeast extract-peptone-dextrose (YPD) and the loss of plasmids was monitored on synthetic complete-His plates. Colonies (clones) were picked and plated on fresh YPD plates. Single colonies were followed thereafter (subclones) by restreaks every 3 days. For the results shown in Fig. 1 and 2, four original 5-FOA-resistant clones were monitored by restreaking each 5-FOA-resistant colony on a fresh YPD plate. Following this first restreak, 24 colonies were picked from each clone and restreaked on fresh YPD plates. Following this second restreak, each of the 96 subclones was followed by serial restreaking of representative colonies, until cessation of cell doubling was observed.
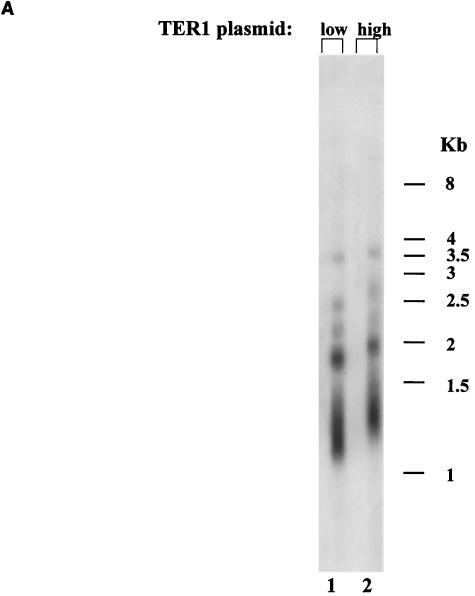
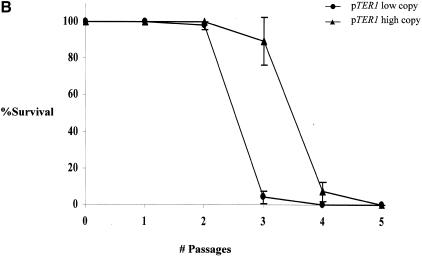
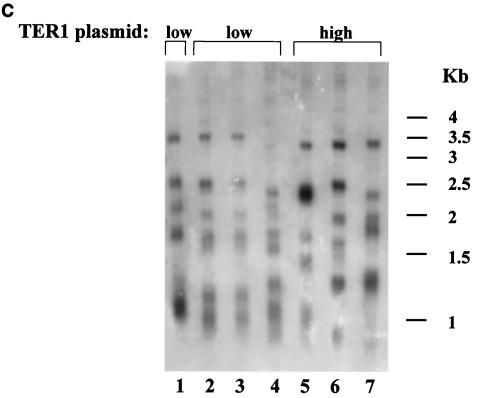
FIG. 1.
Longer telomeres elevate replicative capacity in TER1 K. lactis strains. (A) Genomic DNA was prepared from the indicated strains expressing TER1 from a low-copy-number plasmid (lane 1) or from a high-copy-number plasmid (lane 2). The blot was hybridized with a K. lactis-specific telomeric probe. (B) Percentage of surviving clones with relation to the number of passages in the strain carrying the low- or high-copy-number plasmid. (C) Genomic DNA was prepared from the indicated strains reaching their final cell doublings. Lanes 2 to 4, representative subclones produced from the strain that carried TER1 on a low-copy-number plasmid; lanes 5 to 7, representative subclones produced from the strain that carried TER1 on a high-copy-number plasmid; lane 1, genomic DNA prepared from the strain expressing TER1 from a low-copy-number plasmid prior to loss of the telomerase-bearing plasmid. The blot was hybridized with a K. lactis-specific telomeric probe.
FIG. 2.
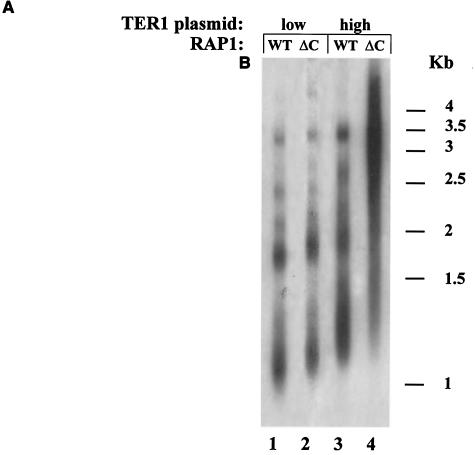
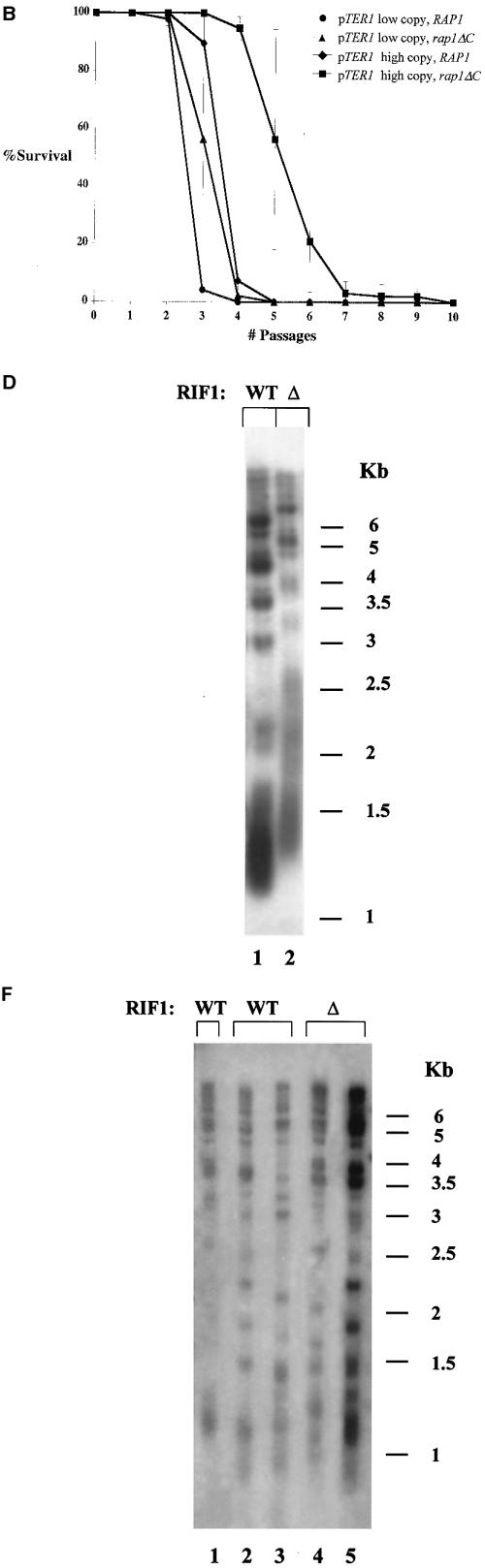
The C-terminal tail of Rap1p in K. lactis and the RIF complex in S. cerevisiae do not function in the determination of replicative capacity. (A) Genomic DNA was prepared from RAP1 (WT) or rap1-ΔC (ΔC) strains expressing TER1 from a low-copy-number plasmid (lanes 1 and 2) or from a high-copy-number plasmid (lanes 3 and 4). The blot was hybridized with a K. lactis-specific telomeric probe. (B) Percentage of surviving clones with relation to the number ofpassages in the RAP1 or rap1-ΔC strain carrying the low- or high-copy-number plasmid. (C) Genomic DNA was prepared from the indicated strains reaching their final cell doublings. Lanes 2 and 3, representative subclones produced from the RAP1 strain that carried TER1 on a low-copy-number plasmid; lanes 4 and 5, representative subclones produced from the rap1-ΔC strain that carried TER1 on a low-copy-number plasmid; lanes 6 and 7, representative subclones produced from the RAP1 strain that carried TER1 on a high-copy-number plasmid; lanes 8 and 9, representative subclones produced from the rap1-ΔC strain that carried TER1 on a high-copy-number plasmid; lane 1, genomic DNA prepared from the RAP1 strain that carried TER1 on a low-copy-number plasmid prior to loss of the telomerase-bearing plasmid. The blot was hybridized with a K. lactis-specific telomeric probe. (D) Genomic DNA was prepared from RIF1 (WT) or Δrif1 (Δ) S. cerevisiae strains. The blot was hybridized with an S. cerevisiae-specific telomeric probe. (E) Percentage of surviving clones with relation to the number of passages in the RIF1 or Δrif1 strain. (F) Genomic DNA was prepared from the RIF1 (lanes 2 and 3) and Δrif1 (lanes 4 and 5) strains reaching their final cell doublings. Lane 1, genomic DNA prepared from the RIF1 strain prior to loss of the telomerase-bearing plasmid. The blot was hybridized with an S. cerevisiae-specific telomeric probe.
For the experiments comparing the replicative capacities of the TER1 and ter1 strains (Fig. 3 and 4) or of the TLC1 and tlc1-C476G strains (Fig. 5), in each strain, three original 5-FOA-resistant clones were restreaked on YPD plates. Following this first restreak, two colonies of each subclone were chosen and again restreaked onto YPD. In this manner, at each passage, every subclone is split into two subclones until eight subclones emanating from a single original 5-FOA-resistant colony are reached (restreak 4). In cases when cessation of cell doublings hasn't been reached at this point, only a single colony was subsequently restreaked from each clone, such that a total of 24 subclones are monitored for each strain.
FIG. 3.
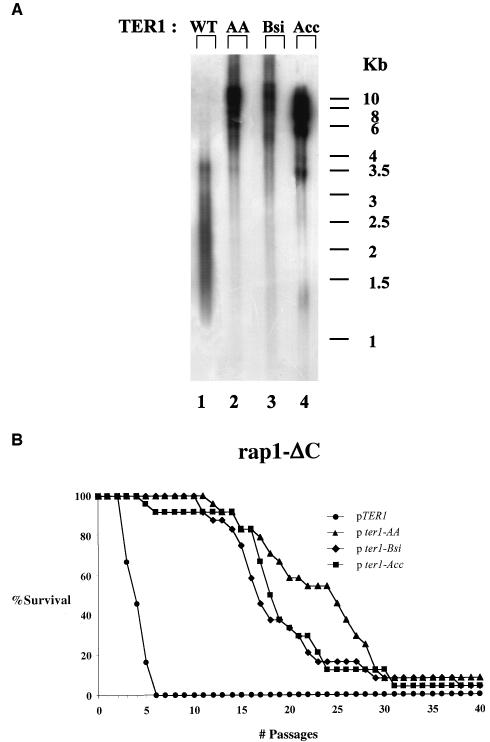
Rap1p binding is not essential for determination of replicative capacity in K. lactis. (A) Genomic DNA was prepared from strains expressing TER1 from a high-copy-number plasmid (WT), ter1-AA (AA), ter1-Bsi (Bsi), or ter1-Acc (Acc). The blot was hybridized with a K. lactis-specific telomeric probe. (B) Percentage of surviving clones with relation to the number of passages in the TER1, ter1-AA, ter1-Bsi, or ter1-Acc strain.
FIG. 4.
The Rap1p-recruited complex is dispensable for determination of replicative capacity in K. lactis. (A) Genomic DNA was prepared from rap1-ΔC strains carrying high-copy-number plasmids expressing TER1 (WT), ter1-AA (AA), ter1-Bsi (Bsi), or ter1-Acc (Acc). The blot was hybridized with a K. lactis-specific telomeric probe. (B) Percentage of surviving clones with relation to the number of passages in the TER1, ter1-AA, ter1-Bsi, or ter1-Acc strain.
FIG. 5.
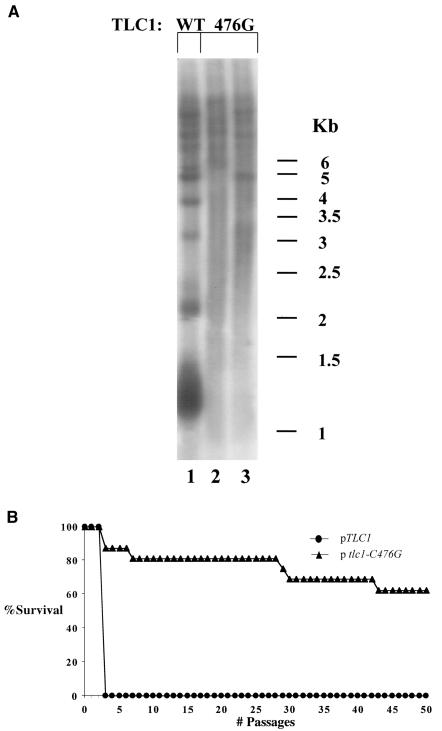
Rap1p binding is not important for determination of replicative capacity in S. cerevisiae. (A) Genomic DNA was prepared from S. cerevisiae strains carrying plasmids expressing TLC11 (WT) or tlc1-C476G (476G). The blot was hybridized with an S. cerevisiae-specific telomeric probe. (B) Percentage of surviving clones with relation to the number of passages in TLC11 or tlc1-C476G.
The final number of restreaks per each subclone was verified by plating serial 10-fold dilutions of cells from colonies in the restreak before the last restreak. All B panel plots show the averages of all the data for every experiment.
For the tables, replicative capacity is calculated for the clones that achieved final capacity. Telomere repeat sizes are calculated by reducing 870 bp of internal nonrepeat sequence for S. cerevisiae, as published previously (34), and 713 bp for K. lactis, as calculated from its telomeric sequence.
Determination of telomere shortening rates.
Colonies were removed from 5-FOA plates upon loss of the respective telomerase-bearing plasmid and placed in 5 ml of liquid YPD. Cultures were allowed to grow to saturation and diluted 1,000-fold.
Four clones of each strain of the wild-type and rap1-ΔC strains were monitored for 4 redilutions (40 cell divisions). From this data, average and standard deviation values were calculated and are presented in the tables.
Since shortening rates of the ter1-Acc strain were very variable between clones and between different telomeres of the same clone, the calculation of shortening rates involved a larger number of measurements. Eight separate liquid cultures were taken for each of the four clones. Shortening rates were calculated for each of these by measuring the difference in telomere length along 35 cell divisions. For each clone, 3 telomeres were chosen for the measurement of telomere shortening. For each initial clone, shortening rates were calculated as the average of the data collected from 3 telomeres of each 8 subclones. From this data, average telomere shortening rates and standard deviation values were calculated and are presented in the tables.
Two-level nested ANOVA statistical test.
Since we observed a high variance in replicative capacity between different clones of each strain, we wanted to determine whether the differences between strains are significant. For this purpose, we used a two-level nested analysis of variance (ANOVA) test.
Two-level nested ANOVA determines whether the variance between subgroups (clones) is different from the variance between groups (strains). The ANOVA test utilizes an F table, specific to the chosen significance (α value). The calculated Fs is tested against the F table. In the table, one should select a value according to the degrees of freedom used in the test (df1, df2).
When testing for significance of difference between strains in their replicative capacity, the Fs should be higher than that in the table (F) at a specific α value, to allow the conclusion that the replicative capacity of clones of the same strain is significantly different than that between clones. A statistical significance of 0.01 (α = 0.01) was chosen. The results are presented as Fs < or > Fα= 0.01(df1, df2).
RESULTS
Telomere length determines the replicative capacity of telomerase-deficient yeast.
To determine which telomeric property determines replicative capacity in the absence of telomerase in the budding yeast K. lactis, we have constructed an experimental system consisting of a deletion of the telomerase RNA gene (TER1) in its chromosomal locus and backed up with a URA3-marked plasmid carrying TER1 or its derivatives. In addition, RAD52, a central gene essential for homologous recombination, was deleted in these strains to avoid the appearance of recombination-dependent survivors in the absence of telomerase (36, 44). By plating these cells on medium containing the uracil analogue 5-FOA, we selected for plasmid loss (cells expressing URA3 are unable to grow in the presence of 5-FOA). These clones are followed thereafter by serial restreaks on YPD plates. The number of restreaks between the loss of the plasmid carrying TER1 and cessation of cell growth of all clones (as observed by inability to form colonies) is defined as the maximal life span of a strain. The number of restreaks needed for 50% of the clones to reach replicative senescence is defined as the average replicative capacity of a strain. In addition, telomere length determination was followed by Southern blotting and hybridization with a telomere-specific probe.
We first wanted to test whether, in our system, replicative capacity indeed correlates with initial telomere length, as expected. To this end, we compared the replicative capacity of two strains that differ only in telomeric lengths. We expected that if initial telomere length is the main determinant of replicative capacity in our system, we would expect that cells starting out with longer telomeres would survive longer than cells which started out with shorter telomeres. In this case, as each strain would stop to divide upon reaching a minimal telomeric size, final telomere lengths are expected to be similar in both strains at the time they reach replicative senescence.
To construct two strains with different telomere lengths, a strain with a deletion of the chromosomal TER1 was transformed with either a low-copy-number plasmid or a high-copy-number plasmid, both of which carried a wild-type copy of the TER1 gene. As shown in Fig. 1A, the telomeres of a strain carrying TER1 on a high-copy-number plasmid (Fig. 1A, lane 2) were longer than those that carried TER1 on a low-copy-number plasmid (Fig. 1A, lane 1) due to a currently unknown mechanism. To compare the replicative capacity of the strains with short and long telomeres, both strains were plated on 5-FOA-containing plates to select for loss of the TER1 gene. For each strain, four original 5-FOA-resistant clones were monitored, by restreaking each 5-FOA-resistant colony on a fresh YPD plate. Following this first restreak, 24 colonies were chosen from each clone and restreaked on fresh YPD plates. Following this second restreak, each of the 96 subclones was followed by serial restreaking of representative colonies, until cessation of cell doubling was observed. In this manner, the replicative capacity of a total of 96 subclones was determined for each strain. As shown in Fig. 1B, the maximal number of restreaks of the strain that started out with longer telomeres, was 4 restreaks (about 100 generations). In comparison, the maximal number of restreaks of the strain starting out with shorter telomeres was 3 restreaks (about 75 generations). The average number of restreaks was 3 for the strain with longer telomeres and 2 for the strain with shorter telomeres [Fs = 106.7 > F0.01 (1, 6) = 13.7]. To determine telomere sizes at the time of replicative senescence, genomic DNA was extracted from cells approaching their final doubling. This was performed by inoculating liquid media with colonies from the last viable streak and incubating them until no additional cell doubling was observed. Exhaustion of replicative capacity was confirmed for each subclone by plating serial dilutions on YPD plates.
The final telomeric lengths were tested for all clones at final cell doublings. Three representative subclones are shown for each strain in Fig. 1C. As seen, although the telomeric patterns of the clones deviated somewhat from each other (Fig. 1C, lanes 2 to 7), the general banding pattern and average sizes of the telomeric fragments at the final cell doublings were similar for all clones. In addition, the shortest fragments that could be detected were of similar sizes in all clones. This may indicate that minimal telomere lengths were similar for all clones tested. The results of this experiment are summarized in Table 1.
TABLE 1.
Telomere length determines the replicative capacity of telomerase-deficient K. lactis
| TER1 strain | Initial telomere length (bp) | Range of final telomere length (bp) | Δ shortening (bp) | Replicative capacity (no. of passages)
|
|
|---|---|---|---|---|---|
| Avga | Maximal | ||||
| Low copy | 495 | 190-215 | 320 | 2.01 ± 0.05 | 3 |
| High copy | 675 | 190-215 | 505 | 3 ± 0.18 | 4 |
Data are means ± standard deviations.
To conclude, in K. lactis, the initial telomere length correlates directly with replicative capacity in the absence of telomere maintaining activities.
The C-terminal tail of Rap1p in K. lactis and the RIF complex in S. cerevisiae are not essential for determination of replicative capacity in the absence of telomerase.
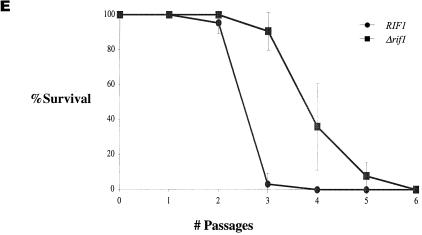
It was previously shown that truncation of the C-terminal tail of Rap1p resulted in a mild increase in telomere length in K. lactis (30) and S. cerevisiae (35). In S. cerevisiae, a larger truncation of the entire C-terminal domain (amino acids 690 to 827) resulted in a drastic increase in telomere length due to the abolishment of binding of two Rap1p-interacting proteins, Rif1p and Rif2p (20, 60). Moreover, it was suggested that a protein counting mechanism regulates telomere length in S. cerevisiae. According to this model, the number of Rap1p carboxy-terminal molecules assembled at the telomere are monitored (43). Theoretically, it was possible that the same mechanism is used to monitor telomere length in the absence of telomerase and thereby determine replicative capacity. If this were the case, we could expect that replicative capacity would decrease in Rap1p mutants lacking telomerase. In contrast, if telomere length were monitored independently of Rap1p, then the initial long telomeres of rap1-ΔC cells in K. lactis or Δrif1 cells in S. cerevisiae would lead to an increased replicative capacity. To distinguish between these possibilities, we compared the replicative capacities of a K. lactis strain carrying a Rap1p C-terminal truncation (rap1-ΔC) with a strain carrying a wild-type copy of RAP1 (Fig. 2A to C). In addition, we compared the replicative capacities of an S. cerevisiae strain carrying a deletion in RIF1 (Δrif1) with a strain carrying a wild-type copy of RIF1 (Fig. 2D to F). The rap1-ΔC allele carries a frameshift mutation at residue 635, producing a polypeptide lacking the normal C-terminal 31 amino acids. For clarity we will refer to this allele as rap1-ΔC. As shown in Fig. 2A, telomere length is elevated in rap1-ΔC compared to RAP1 in strains expressing TER1 from a low-copy-number plasmid (Fig. 2A, compare lane 2 to lane 1) and in strains expressing TER1 from a high-copy-number plasmid (Fig. 2A, compare lane 4 to lane 3), which is as expected. As shown in Fig. 2B, truncation of the C-terminal tail of Rap1p did not reduce the replicative capacity of cells in the absence of telomerase. In contrast, the strain expressing TER1 from a high-copy-number plasmid in the background of a C-terminal truncation of Rap1p, had acquired a significantly higher replicative capacity (average of 4.8 passages) than a strain expressing TER1 from a multicopy plasmid in a RAP1 background (average of 3 passages) [Fs = 26.6 > F0.01 (1, 6) = 13.7]. Again, telomere lengths at the time of exhaustion of cell doubling capacity were tested for all clones and, as shown for two representative clones of each strain, were similar in all strains tested in this experiment, regardless of their initial lengths (Fig. 2C). Again, as seen in the previous experiment, all clones had fragments that migrated faster than 1 kb, and here too, telomeric patterns differed somewhat from one clone to another.
We conclude that initial telomere length, regardless of the presence or absence of the C-terminal tail domain of Rap1p, determines the replicative capacity of K. lactis in the absence of telomerase.
Since these experiments were performed in K. lactis, where the role of RIF proteins has not been investigated yet and a larger deletion of the C-terminal domain of Rap1p is lethal (our unpublished results), we turned to S. cerevisiae in order to investigate directly the role of the RAP1-RIF complex in the determination of replicative capacity in the absence of telomerase. To this end, we have created a Δrif1 strain carrying a deletion of the chromosomal copy of the telomerase RNA TLC1 gene and a plasmid carrying TLC1 and the URA3 gene.
As shown in Fig. 2D and previously published, telomere length is elevated in a Δrif1 strain (Fig. 2D, lane 2) compared to a RIF1 strain (Fig. 2D, lane 1) (both strains also had deletions of RAD52). As shown in Fig. 2E, deletion of RIF1 did not reduce the replicative capacity of cells in the absence of telomerase. In contrast, the strain carrying a deletion of this gene had acquired a significantly higher replicative capacity (average of 3.4 passages) than a RIF1 strain (average of 1.9 passages) [Fs = 90.7 > F0.01 (1, 6) = 13.7]. Again, telomere lengths at the time of exhaustion of cell doubling capacity were tested for all clones and, as shown for two representative clones of each strain, were similar in a RIF1 strain (Fig. 2E, lanes 2 and 3) and a Δrif1 strain (Fig. 2E, lanes 4 and 5), regardless of their initial lengths (Fig. 2F). All clones had fragments that migrated faster than 1.2 kb, and here too, telomeric patterns differed somewhat from one clone to another.
Therefore, replicative capacity was directly correlated to initial telomere length in the presence or absence of Rif1p. The results of these experiments are summarized in Table 2.
TABLE 2.
The C-terminal tail of Rap1p is not essential for determination of replicative capacity of telomerase-deficient K. lactis, and RIF1 is not essential for determination of replicative capacity of telomerase-deficient S. cerevisiae
| Strain | Initial telomere length (bp) | Range of final telomere length (bp) | Δ shortening (bp) | Replicative capacity (no. of passages)
|
|
|---|---|---|---|---|---|
| Avga | Maximal | ||||
| RAP1 low copy | 495 | 190-215 | 320 | 2.01 ± 0.05 | 3 |
| rap1-ΔC low copy | 525 | 190-215 | 335 | 2.58 ± 0.18 | 4 |
| RAP1 high copy | 675 | 190-215 | 505 | 3 ± 0.18 | 4 |
| rap1-ΔC high copy | 1,325 | 210-340 | 1,110 | 4.79 ± 0.68 | 9 |
| RIF1 (S. cerevisiae) | 500 | 205-245 | 295 | 1.91 ± 0.13 | 3 |
| Δrif1 (S. cerevisiae) | 1,100 | 220-340 | 790 | 3.38 ± 0.27 | 5 |
Data are means ± standard deviations.
Rap1p binding potential is not critical for determination of replicative capacity in telomerase-deficient K. lactis cells.
Mutations in the template region of the telomerase RNA (TER1) gene direct the incorporation of the altered telomeric DNA sequence (45). It was previously shown that incorporation of base substitutions, which disrupted the Rap1p binding site, resulted in the loss of telomere length control (45). The phenotypic severity of these mutations in vivo correlates directly with the degree of loss of Rap1p binding affinity to the mutated telomeric sequence in vitro (30). If telomere length per se determines replicative capacity in the absence of telomerase, strains initiating with mutant ter1 and, therefore, with longer telomeres are expected to have an extended replicative capacity relative to a strain initiating with shorter telomeres. In contrast, if the reduction in Rap1p binding throughout the telomere affects its structure in a way that accelerates cellular senescence, we would expect that the replicative capacity of strains initiating with long telomeres due to reduced Rap1p binding potential would be reduced compared to that of strains with normally complexed telomeres. We would also expect that ter1 mutants would cease to divide while their telomeres are longer than those of wild-type cells. This is due to the fact that those longer telomeres would in fact be measured as being short if what is being measured is bound Rap1p and not actual DNA length. It was previously observed with a K. lactis strain with mildly elongated AA telomeres that, in the presence of RAD52, changes in colony morphology occurred later than in a strain carrying normal repeats. However, maximal and average replicative capacities were not tested in this system (44).
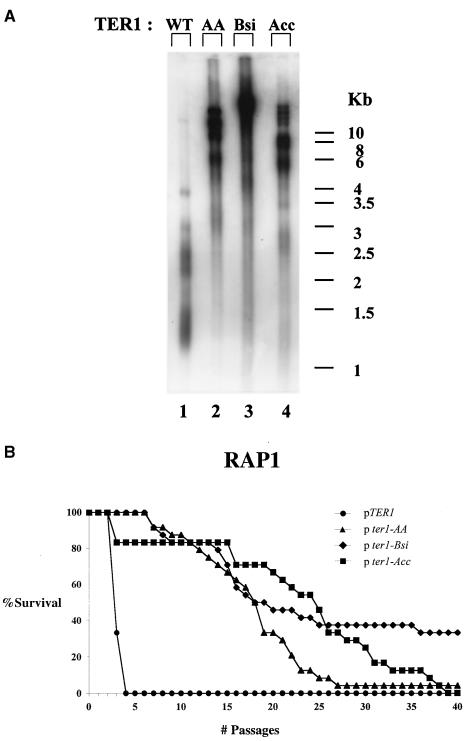
To study the effect of Rap1p binding potential throughout the telomere on the replicative capacity of yeast in the absence of telomerase, we compared the replicative capacities of strains carrying either wild-type telomeric repeats (TER1 strain) or mutated repeats, AA, Bsi, or Acc (the ter1-AA, ter1-Bsi, or ter1-Acc strain, respectively). All strains had a deletion of RAD52.
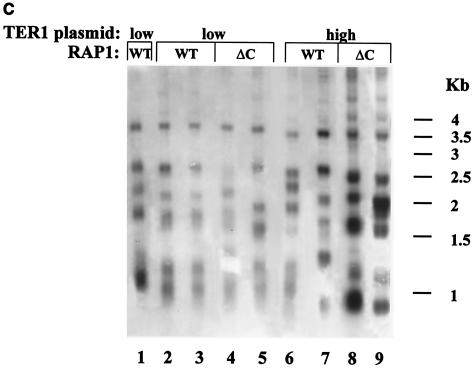
It has previously been shown that these mutations led to 10-, 100-, and 300-fold reductions, respectively, in Rap1p binding potential in vitro and had lost telomere length control in vivo (30). As shown in Fig. 3A, telomeres of ter1-AA, ter1-Bsi, and ter1-Acc strains (Fig. 3A, lanes 2, 3, and 4, respectively) are much longer than those of TER1 (Fig. 3A, lane 1), due to reduced Rap1p binding to the telomeres of these strains. Moreover, the telomeric banding pattern became very heterogeneous, as judged by the smeary appearance of the telomere-reactive signal, indicative of loss of telomere length regulation. This is in agreement with previously published data (45, 48). The telomeres of each ter1 mutant strain are composed of a short basal section of wild-type repeats and a very long terminal section of mutant repeats that were added upon it. It should be noted that in the current system, telomere lengths seem similar in the three ter1 mutant strains tested. This is probably the result of overexpression of the mutant telomerase RNAs, which apparently synergizes with the reduced Rap1p binding with respect to loss of telomere length control. As seen in Fig. 3B, the average and maximal replicative capacity of either mutant ter1 strain (p ter1 strains) was greatly increased (over 7- and 10-fold) compared to the TER1 strain (pTER1 strain) [among strains, for ter1-AA, Fs = 123.5 > F0.01 (1,4) = 21.2; for ter1-Bsi, Fs = 47 > F0.01 (1,4) = 21.2; for ter1-Acc, Fs = 60.1 > F0.01 (1,4) = 21.2] [among clones of each strain, for ter1-AA, Fs = 0.6 < F0.01 (4,41) = 3.8; for ter1-Bsi, Fs = 0.43 < F0.01 (4,41) = 3.8; for ter1-Acc, Fs = 1 < F0.01 (4,41) = 3.8]. The results of this experiment are summarized in Table 3.
TABLE 3.
Rap1p binding potential is not critical for determination of replicative capacity of telomerase-deficient K. lactis strains
| Strain | Initial telomere length (kb) | Range of final telomere length (kb) | Δ shortening (kb) | Replicative capacity (no. of passages)
|
|
|---|---|---|---|---|---|
| Avgd | Maximal | ||||
| TER1 | 0.49 | 0.19-0.21 | 0.32 | 3 ± 0.18 | 4 |
| ter1-AA | 6-10a | 0.25-10a | 5.75-9.75b | 16.1 ± 2.2 | 40 |
| ter1-Bsi | 6-10a | 0.25-10a | 5.75-9.75b | 20.9 ± 12.1c | 40+ |
| ter1-Acc | 6-10a | 0.25-10a | 5.75-9.75b | 21 ± 4.2c | 39 |
The upper limit of the telomeric bands is at the limit of mobility of the gels, around 10 kb.
It is impossible to define the source of the fastest migrating fragments; therefore, the Δ shortening is shown for either possibility (fastest migrating fragment or slowest migrating initial fragment shortening to the fastest migrating fragments).
Calculated only for those clones that achieved final replicative capacity.
Data are means ± standard deviations.
It should be noted that we have not seen any consistent differences between clones that emerged from different founder colonies. Differences between the strains were significantly larger than differences between clones of the same strain. Additionally, it should be noted that, especially for the ter1-bsi strain, replicative capacity did not follow a binomial distribution. Instead, there was a leveling off of survival due to the fact that some of the clones monitored did not lose replicative capacity during the course of the experiment. This may be due to some mode of survival that is not dependent on homologous recombination (since all strains tested were Δrad52). In some of these clones, a banding pattern could be observed in the absence of telomerase, which remained steady during many restreaks (data not shown). This may indicate that fusions between telomeres had occurred. Such fusions may stop telomere shortening and therefore account for the change in distribution.
To summarize this experiment, reduced Rap1p binding potential, which results in deregulation of telomere length, leads to a dramatic increase in the replicative capacity of cells deficient for telomerase. We conclude that telomere length per se, and not Rap1p occupancy throughout the telomere, determines replicative capacity in the absence of telomerase.
The replicative capacity of telomerase-deficient K. lactis cells with reduced Rap1p binding potential is not affected by a truncation of the C-terminal tail of Rap1p.
As mentioned, we have previously seen that disruption of Rap1p binding to the telomere and truncation of Rap1p C terminus act synergistically to deregulate telomere length. We wanted to test whether these double mutations also act synergistically to affect the replicative capacity of telomerase-deficient cells. To this aim, we followed the fate of the double mutants ter1-AA rap1p-ΔC, ter1-Bsi rap1p-ΔC, and ter1-Acc rap1p-ΔC upon loss of telomerase. All strains had deletions of RAD52. As shown in Fig. 4A, lanes 2 to 4, telomeres of the double mutants were considerably longer than those of the single rap1p-ΔC mutant (Fig. 4A, lane 1).
Figure 4B shows that the average and maximal replicative capacity of either double mutant strain (pter1 strains) was greatly increased (over four- and fivefold) compared to that of the single rap1p-ΔC strain (pTER1 strain) [among strains, for ter1-AA, Fs = 54.3 > F0.01 (1,4) = 21.2; for ter1-Bsi, Fs = 197.6 > F0.01 (1,4) = 21.2; for ter1-Acc, Fs = 4649.5 > F0.01 (1,4) = 21.2] [among clones of each strain, for ter1-AA, Fs = 1.3 < F0.01 (4,41) = 3.8; for ter1-Bsi, Fs = 0.2 < F0.01 (4,41) = 3.8; for ter1-Acc, Fs = 0.01 < F0.01 (4,41) = 3.8]. The results of this experiment are summarized in Table 4.
TABLE 4.
Replicative capacity of telomerase-deficient K. lactis strains with reduced Rap1p binding potential is not affected by truncation of the C-terminal tail of Rap1p
| Strain | Initial telomere length (kb) | Range of final telomere length (kb) | Δ shortening (bp) | Replicative capacity (no. of passages)
|
|
|---|---|---|---|---|---|
| Avgc | Maximal | ||||
| rap1-ΔC TER1 | 0.675 | 0.19-0.21 | 505 | 4.79 ± 0.68 | 9 |
| rap1-ΔC ter1-AA | 6-10+a | 0.25-10+a | 5.75-9.75b | 21.6 ± 4.2 | 40 |
| rap1-ΔC ter1-Bsi | 6-10+a | 0.25-10+a | 5.75-9.75b | 18 ± 2 | 40 |
| rap1-ΔC ter1-Acc | 6-10+a | 0.25-10+a | 5.75-9.75b | 17.9 ± 0.37 | 39 |
The upper limit of the telomeric bands is at the limit of mobility of the gels, around 10 kb.
It is impossible to define the source of the fastest migrating fragments; therefore, the Δ shortening is shown for either possibility (fastest migrating fragment or slowest migrating initial fragment shortening to the fastest migrating fragments).
Data are means ± standard deviations.
To conclude, elevated telomere length, regardless of Rap1p binding potential and in spite of the Rap1p C-terminal tail truncation, leads to a dramatic increase in the replicative capacity of cells correlating with their effects on telomere length.
Rap1p binding potential is not critical for determination of replicative capacity in telomerase-deficient S. cerevisiae cells.
Many properties of the telomere length regulation machinery are shared between S. cerevisiae and K. lactis. Among these is the effect of disruption of Rap1p binding to the telomere. In S. cerevisiae, a mutation in the template domain of telomerase RNA, TLC1, termed tlc1-C476G, which reduces Rap1p in vitro binding affinity to the telomeric repeats by 300-fold, results in an increase in telomere length in a manner similar to that observed in K. lactis ter1-Acc (51) (Fig. 5, compare lanes 2 and 3 to lane 1). To study whether the role of telomere length in the determination of replicative capacity in the absence of telomerase is unique to K. lactis or shared with S. cerevisiae, we compared the replicative capacity of a strain carrying normal telomeres to that of a strain carrying the tlc1-C476G mutation following loss of telomerase. All strains had deletions of RAD52. As shown in Fig. 5B, following loss of telomerase, the maximal replicative capacity of a TLC1 strain was 2 restreaks while that of the tlc1-476G strain was dramatically increased and at the time of this publication over 50% of the clones followed still continued beyond 80 passages [Fs = 674.9 > F0.01 (1, 2) = 98.5]. Surviving clones were not included in the statistical calculations, since they must have arisen due to some RAD52-independent mode of survival. The results of this experiment are summarized in Table 5.
TABLE 5.
Rap1p binding potential is not critical for determination of the replicative capacity of telomerase-deficient S. cerevisiae strains
| Strain | Initial telomere length (kb) | Range of final telomere length (kb) | Δ shortening (bp) | Replicative capacity (no. of passages)
|
|
|---|---|---|---|---|---|
| Avgd | Maximal | ||||
| TLC1 | 0.54 | 0.21-0.24 | 295 | 1.91 ± 0.31 | 2 |
| tlc1-C476G | 6-10+a | 6-10+a | NDb | 23.3 ± 3.8c | 80+ |
The upper limit of the telomeric bands is at the limit of mobility of the gels, around 10 kb.
Shortening cannot be seen at the gels' resolution.
Calculated only for those clones that achieved final replicative capacity.
Data are means ± standard deviations.
To conclude, in S. cerevisiae, as in K. lactis, the reduction in Rap1p binding to the telomere, resulting in the elongation of telomeres, leads to an increase in replicative capacity.
DISCUSSION
Longer telomeres extend replicative capacity in the absence of telomere maintenance activities.
The limitation of replicative capacity shown for organisms from ciliates and yeasts to human primary cells in culture was suggested to be an important factor in aging, and considered a major obstacle for uncontrolled cell growth and tumor progression.
In our study of the specific telomere properties and the mechanism by which cell division is arrested in budding yeasts lacking telomerase and alternative telomere lengthening activities, we have set out to study the telomere properties that determine replicative capacity by measuring the number of cell divisions as a function of initial telomere length.
The main finding of this work is that in the absence of telomerase, the initial length of telomeres determines the number of cell doublings a yeast cell can undergo before it exhausts its replicative capacity, regardless of the presence of the Rap1p C-terminal domain, a Rap1p-recruited complex, and even Rap1p binding. Furthermore, a gradient of replicative capacity is obtained which follows closely a gradient of initial telomere length.
Nonessential telomeric properties.
In S. cerevisiae, it was seen that, in vitro, telomerase remains associated with its substrate (50). In addition, a protective function was indicated for human telomerase (61). The close correlation between telomere length and replicative capacity clearly shows that the absence of telomerase by itself does not determine replicative capacity.
The binding of Rap1p and its associated protein complex was shown to play a central role in many telomere-associated functions, including protein counting of the number of telomere-bound Rap1p molecules and their interacting factors, which measures telomere length (43) and is essential for telomeric position effect (10, 38, 40) and meiosis (8, 41). It was therefore possible that in the absence of telomerase, the number of Rap1p molecules and/or Rap1p-associated factors bound throughout the entire telomere are counted by a similar mechanism and determine replicative capacity. However, this work clearly shows that K. lactis and S. cerevisiae strains whose telomeres are extremely long due to their reduced ability to bind Rap1p (with either wild-type or C-terminally truncated Rap1p and with or without the RAP-RIF complex, respectively) had the most extended replicative capacities.
Furthermore, the similar replicative capacities of the ter1-AA and ter1-Acc strains, despite the expected 30-fold difference in their ability to bind Rap1p, show that the extent of Rap1p binding throughout the telomere is not a critical factor in determining replicative capacity. From this we conclude that the binding of Rap1p throughout the telomere and the nucleation of the putative Rap1p-recruited protein complex are not central for replicative capacity.
It was suggested that changes in the amount of single-stranded telomeric DNA might be the cause for replicative senescence in certain situations. We have tested the amount of single-stranded DNA in our systems (data not shown) and found that in K. lactis there is a small amount of single-stranded telomeric DNA in the wild-type strain and this amount does not change throughout the course of the experiment. As for the ter1-Acc strain, there is a very large amount of single-stranded telomeric DNA, even in the presence of telomerase, which remains similar throughout most of the experiment. Therefore, a large amount of single-stranded telomeric DNA cannot account for the loss of replicative capacity in our system. In S. cerevisiae, both wild-type and tlc1-476 strains had a small amount of single-stranded telomeric DNA in the presence of telomerase and this amount did not change during the course of the experiment. Thus, accumulation or loss of single-stranded telomeric DNA cannot account for the loss of replicative capacity in our systems.
Interestingly, our results show that the reduction in binding of telomeres by Rap1p enhances the shortening of telomeres in the absence of telomere maintenance activities. Initial telomere lengths are estimated to be over 100 times longer in ter1 strains than in the wild-type strain (41, 45); however, the maximal number of restreaks is only 10-fold greater (Table 4, compare TER1 and ter1-Acc). We have calculated the rate of telomere shortening in the various strains by following consecutive restreaks in liquid media, as shown in Table 6. The main conclusion from this table is that the rate of telomere shortening is indeed enhanced (1.5-fold, minimally) in ter1-Acc and in the rap1-ΔC strain, which had TER1 on a high-copy-number plasmid. It is possible that when telomeres are above a certain length, their shortening rate is enhanced following the loss of telomerase. Alternatively, it is possible that some other factor besides telomere length affects telomere shortening, such as telomere heterogeneity. According to this model, when telomeres of the same cell are of different sizes, as in the strains tested, some telomere shortening activity is enhanced, leading to enhancement of shortening, thereby reducing the replicative capacity.
TABLE 6.
Telomere shortening rates of K. lactis strainsa
| Strain | Shortening rate (bp/division) |
|---|---|
| RAP1 TER1 | 5.65 ± 0.94 |
| rap1-ΔC TER1 | 8.83 ± 1.67 |
| RAP1 ter1-Acc | 8.92 ± 3.93 |
Four clones were taken for each experiment. Telomere shortening was calculated (from 4 to 8 measurements for each point) as the difference in telomere length between the first and fifth dilutions. TER1 and ter1-Acc were high-copy-number plasmids. Data are means ± standard deviations.
Alternatively, changes in the telomeric protein complex affect the rate of telomere shortening. In S. cerevisiae, it was previously shown that the rate of telomere shortening in the absence of telomerase was enhanced twofold in mutants lacking the entire C-terminal domain of Rap1p (42).
How is telomeric length counted?
Our work clearly shows that a minimal length of telomeric DNA is essential for the continuation of cell division. There are a few possibilities for envisioning how this length is monitored. In mammalian cells, TRF2 is needed for the formation in vitro of a special terminal structure termed the t-loop (18, 57). T-loops were also found at the ends of micronuclear Trypanosoma brucei telomeres (49). It is possible that in yeast a minimum length of double-stranded telomeric DNA is needed for the formation of a yet unidentified structure which is necessary for capping the chromosome end and thereby for the continuation of cell division. The clustering of telomeres at limited domains of the cell nucleus during meiosis and mitosis (13, 16, 53), known as telomere clustering, has been observed in a wide variety of organisms. This cytological phenomenon may reflect the existence of functional telomeric domains. It is possible that critically shortened telomeres are unable to participate in such intertelomeric interactions and are therefore nonfunctional. Alternatively, shortened telomeres may not be able to participate in correct nuclear positioning (15, 24).
Another possibility for the eventual cessation of cell doubling is that the binding of a protein other than Rap1p is necessary to protect telomeres. This putative protein must be able to bind telomeres of TER1 (and TLC1) as well as those of ter1 (and the tlc1-C476G strain). Once the number of these proteins associated with telomeres falls short of a minimal number due to the critical shortening of telomeres, cell division stops. This may be similar to the proposed role of TRF2 in mammalian primary cells (29). The existence of a Rap1p-independent counting mechanism has been recently proposed (7).
It is still possible that a minimal number of internal, rather than terminal, Rap1p-bound telomeric repeats is counted. In this case, Rap1p molecules (regardless of the existence of the C-terminal tail) are counted only once the telomeric DNA associated with them becomes terminal.
How do short telomeres lead to senescence?
In mammalian cells, the expression of dominant-negative TRF2 and telomere shortening in mice lacking telomerase result in p53-dependent apoptosis and cell cycle arrest (28). It is possible that in yeast a checkpoint system exists that monitors and signals telomeric size, leading to an active cell cycle arrest when telomeres are too short, as suggested previously (14, 27). Alternatively, a critically short telomere may lead to cessation of cell doubling by engaging in illegitimate chromosome fusions due to loss of capping, leading to catastrophic chromosomal aberrations, or it may lose its ability to perform some function which is essential for cell viability.
Acknowledgments
We thank Martin Kupiec, Dudy Tzfati, and Titia de Lange for helpful comments on the manuscript. We also thank John Prescott for the tlc1-C476G plasmid and Michael McEachern for the K. lactis RAD52 deletion strain.
This work was supported by grants from the Israel Cancer Research Fund and The United States-Israel Binational Science Fund.
Footnotes
Dedicated to the children of Anat Krauskopf, Yotam and Merav.
REFERENCES
- 1.Allsopp, R. C., E. Chang, M. Kashefi-Aazam, E. I. Rogaev, M. A. Piatyszek, J. W. Shay, and C. B. Harley. 1995. Telomere shortening is associated with cell division in vitro and in vivo. Exp. Cell Res. 220:194-200. [DOI] [PubMed] [Google Scholar]
- 2.Allsopp, R. C., H. Vaziri, C. Patterson, S. Goldstein, E. V. Younglai, A. B. Futcher, C. W. Greider, and C. B. Harley. 1992. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. USA 89:10114-10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackburn, E. H. 1984. The molecular structure of centromeres and telomeres. Annu. Rev. Biochem. 53:163-194. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn, E. H. 1994. Telomeres: no end in sight. Cell 77:621-623. [DOI] [PubMed] [Google Scholar]
- 5.Blasco, M. A., H. W. Lee, M. P. Hande, E. Samper, P. M. Lansdorp, R. A. DePinho, and C. W. Greider. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25-34. [DOI] [PubMed] [Google Scholar]
- 6.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 7.Brevet, V., A. S. Berthiau, L. Civitelli, P. Donini, V. Schramke, V. Geli, F. Ascenzioni, and E. Gilson. 2003. The number of vertebrate repeats can be regulated at yeast telomeres by Rap1-independent mechanisms. EMBO J. 22:1697-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chikashige, Y., and Y. Hiraoka. 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr. Biol. 11:1618-1623. [DOI] [PubMed] [Google Scholar]
- 9.Chin, L., S. E. Artandi, Q. Shen, A. Tam, S. L. Lee, G. J. Gottlieb, C. W. Greider, and R. A. DePinho. 1999. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97:527-538. [DOI] [PubMed] [Google Scholar]
- 10.Cockell, M., F. Palladino, T. Laroche, G. Kyrion, C. Liu, A. J. Lustig, and S. M. Gasser. 1995. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J. Cell Biol. 129:909-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper, J. P., E. R. Nimmo, R. C. Allshire, and T. R. Cech. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385:744-747. [DOI] [PubMed] [Google Scholar]
- 12.Counter, C. M. 1996. The roles of telomeres and telomerase in cell life span. Mutat. Res. 366:45-63. [DOI] [PubMed] [Google Scholar]
- 13.Dernberg, A. F., J. W. Sedat, W. Z. Cande, and H. W. Bass. 1995. Cytology of telomeres, p. 96-105. In E. Blackburn and C. Greider (ed.), Telomeres. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.Enomoto, S., L. Glowczewski, and J. Berman. 2002. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2626-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feuerbach, F., V. Galy, E. Trelles-Sticken, M. Fromont-Racine, A. Jacquier, E. Gilson, J. C. Olivo-Marin, H. Scherthan, and U. Nehrbass. 2002. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 4:214-221. [DOI] [PubMed] [Google Scholar]
- 16.Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Scherthan, and S. M. Gasser. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greider, C. W., and E. H. Blackburn. 1989. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337:331-337. [DOI] [PubMed] [Google Scholar]
- 18.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 19.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 20.Hardy, C. F., L. Sussel, and D. Shore. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6:801-814. [DOI] [PubMed] [Google Scholar]
- 21.Harley, C. B. 1991. Telomere loss: mitotic clock or genetic time bomb? Mutat. Res. 256:271-282. [DOI] [PubMed] [Google Scholar]
- 22.Harley, C. B., A. B. Futcher, and C. W. Greider. 1990. Telomeres shorten during ageing of human fibroblasts. Nature 345:458-460. [DOI] [PubMed] [Google Scholar]
- 23.Hayflick, L. 1965. The limited in vitro lifetime of human diploid cell strains. Exp. Cell Res. 37:614-636. [DOI] [PubMed] [Google Scholar]
- 24.Hediger, F., and S. M. Gasser. 2002. Nuclear organization and silencing: putting things in their place. Nat. Cell Biol. 4:E53-E55. [DOI] [PubMed] [Google Scholar]
- 25.Hemann, M. T., K. L. Rudolph, M. A. Strong, R. A. DePinho, L. Chin, and C. W. Greider. 2001. Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol. Biol. Cell 12:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemann, M. T., M. A. Strong, L. Y. Hao, and C. W. Greider. 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:67-77. [DOI] [PubMed] [Google Scholar]
- 27.IJpma, A. S., and C. W. Greider. 2003. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14:987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlseder, J., D. Broccoli, Y. Dai, S. Hardy, and T. de Lange. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283:1321-1325. [DOI] [PubMed] [Google Scholar]
- 29.Karlseder, J., A. Smogorzewska, and T. de Lange. 2002. Senescence induced by altered telomere state, not telomere loss. Science 295:2446-2449. [DOI] [PubMed] [Google Scholar]
- 30.Krauskopf, A., and E. H. Blackburn. 1996. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383:354-357. [DOI] [PubMed] [Google Scholar]
- 31.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, H. W., M. A. Blasco, G. J. Gottlieb, J. W. N. Horner, C. W. Greider, and R. A. DePinho. 1998. Essential role of mouse telomerase in highly proliferative organs. Nature 392:569-574. [DOI] [PubMed] [Google Scholar]
- 33.Lingner, J., T. R. Cech, T. R. Hughes, and V. Lundblad. 1997. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. USA 94:11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linskens, M. H., C. B. Harley, M. D. West, J. Campisi, and L. Hayflick. 1995. Replicative senescence and cell death. Science 267:17. [DOI] [PubMed] [Google Scholar]
- 35.Liu, C., X. Mao, and A. J. Lustig. 1994. Mutational analysis defines a C-terminal tail domain of RAP1 essential for telomeric silencing in Saccharomyces cerevisiae. Genetics 138:1025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 37.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 38.Luo, K., M. A. Vega-Palas, and M. Grunstein. 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16:1528-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lustig, A. J., S. Kurtz, and D. Shore. 1990. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250:549-553. [DOI] [PubMed] [Google Scholar]
- 40.Lustig, A. J., C. Liu, C. Zhang, and J. P. Hanish. 1996. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2483-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maddar, H., N. Ratzkovsky, and A. Krauskopf. 2001. Role for telomere cap structure in meiosis. Mol. Biol. Cell 12:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcand, S., V. Brevet, and E. Gilson. 1999. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 44.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822-1834. [DOI] [PubMed] [Google Scholar]
- 45.McEachern, M. J., and E. H. Blackburn. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376:403-409. [DOI] [PubMed] [Google Scholar]
- 46.McEachern, M. J., and S. Iyer. 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7:695-704. [DOI] [PubMed] [Google Scholar]
- 47.McEachern, M. J., A. Krauskopf, and E. H. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 48.McEachern, M. J., D. H. Underwood, and E. H. Blackburn. 2002. Dynamics of telomeric DNA turnover in yeast. Genetics 160:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munoz-Jordan, J. L., G. A. Cross, T. de Lange, and J. D. Griffith. 2001. t-loops at trypanosome telomeres. EMBO J. 20:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prescott, J., and E. H. Blackburn. 1997. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 11:2790-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prescott, J. C., and E. H. Blackburn. 2000. Telomerase RNA template mutations reveal sequence-specific requirements for the activation and repression of telomerase action at telomeres. Mol. Cell. Biol. 20:2941-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 53.Scherthan, H. 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2:621-627. [DOI] [PubMed] [Google Scholar]
- 54.Shippen-Lentz, D., and E. H. Blackburn. 1990. Functional evidence for an RNA template in telomerase. Science 247:546-552. [DOI] [PubMed] [Google Scholar]
- 55.Shore, D. 1997. Telomerase and telomere-binding proteins: controlling the endgame. Trends Biochem. Sci. 22:233-235. [DOI] [PubMed] [Google Scholar]
- 56.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 57.Stansel, R., T. de Lange, and J. D. Griffith. 2001. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 20:5532-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stewart, S. A., I. Ben-Porath, V. J. Carey, B. F. O'Connor, W. C. Hahn, and R. A. Weinberg. 2003. Erosion of the telomeric single-strand overhang at replicative senescence. Nat. Genet. 33:492-496. [DOI] [PubMed] [Google Scholar]
- 59.van Steensel, B., and T. de Lange. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385:740-743. [DOI] [PubMed] [Google Scholar]
- 60.Wotton, D., and D. Shore. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11:748-760. [DOI] [PubMed] [Google Scholar]
- 61.Zhu, J., H. Wang, J. M. Bishop, and E. H. Blackburn. 1999. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc. Natl. Acad. Sci. USA 96:3723-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]