Abstract
Intercellular communication plays a key role in the regulation of several physiological processes in gram-positive bacteria. Cell-cell communication is often mediated by secreted inducer peptide pheromones (IPs), which upon reaching a threshold concentration in the environment specifically activate a cognate membrane-localized histidine protein kinase (HPK). Interestingly, the majority of IP-activated HPKs fall into one distinct subfamily (HPK10). As part of an effort to study the mechanism underlying pheromone-mediated activation of the HPK10 subfamily, the present work investigated the membrane topology of PlnB from Lactobacillus plantarum. Gene fusion experiments with Escherichia coli and Lactobacillus sakei, using alkaline phosphatase, β-lactamase, and β-galactosidase reporter fusions, suggested that PlnB is anchored to the cytoplasmic membrane via seven transmembrane segments. By domain switching between HPK10 members, it was demonstrated that the determinants for pheromone binding and specificity are contained within the transmembrane domain. The results also indicate that the mechanism of signal transduction, in which the final transmembrane segment apparently plays a key role, is conserved between members of the HPK10 subfamily.
During the last decade it has become evident that gram-positive bacteria often utilize peptide-inducible two-component signal transduction systems for intercellular communication. Such group behavior has been shown to regulate a diverse set of processes in gram-positive bacteria, including bacteriocin production, natural genetic transformation, and virulence (10, 12, 17, 18, 30, 31, 35). In all of these systems, the signaling molecule perceived by the histidine protein kinase (HPK) sensor is a so-called inducer peptide pheromone (IP) produced by the bacteria themselves. In a recent review, it was revealed that histidine kinases could be divided into distinct subfamilies based on their degree of amino acid homology in the kinase domain. Interestingly, it was shown that all known peptide pheromone-activated HPKs except SpaK, ComP, and NisK fall into the same subfamily (HPK10) (13). The HPK10 subfamily includes, among others, VirS from Clostridium (26), PlnB from Lactobacillus (5), ComD from Streptococcus (16), AgrC from Staphylococcus (14, 34), and CbnK from Carnobacterium (38). All members of the HPK10 subfamily belong to the orthodox kinases, which are constructed of a membrane-spanning N-terminal domain and a C-terminal cytoplasmic kinase domain (13, 47). However, they differ from most membrane-localized histidine kinases in two respects. First, histidine kinase core domains usually contain a set of conserved regions (the N, D, F, and G boxes) that are involved in nucleotide binding. However, the HPK10 nucleotide binding domain apparently lacks a D box and contains only one asparagine in the N box (13). Second, in the majority of prokaryotic HPKs, the N-terminal membrane-associated domain consists of two transmembrane segments (TMS) flanking an extracytoplasmatic loop (33, 47). In contrast, the HPK10 subfamily is predicted to posses a membrane domain containing six or seven TMS (5, 16, 18, 25).
With the exception of the thiolactone-containing IP AgrD from Staphylococcus aureus, all characterized IPs that activate kinases from the HPK10 subfamily are unmodified peptides. They consist of 14 to 27 residues and are usually synthesized as precursor peptides with an N-terminal Gly-Gly leader (23). The double-glycine leader functions as a secretion signal that is removed concomitant with export by a dedicated ABC transporter (15). The different IPs share some striking similarities in that they are cationic molecules which probably adopt α-helical structures in membrane environments (19, 23). However, they share low sequence identity and activate their cognate HPK in a highly specific manner (3, 6, 16, 32, 37).
One of the best-characterized peptide-inducible two-component systems utilizing a HPK from the HPK10 subfamily is the PlnABCD system, which regulates the bacteriocin production of Lactobacillus plantarum C11. In this system, it has been experimentally demonstrated that the PlnA IP (IP-C11) activates the PlnB HPK, thereby triggering a phosphotransfer pathway leading to phosphorylation and subsequent activation of the two response regulators PlnC and PlnD. These proteins function as transcription factors, regulating transcription from the bacteriocin operons (plnEFI and plnJKLR) as well as the regulatory operon itself (plnABCD) (6, 7, 8, 9, 19, 39, 40). In an effort to determine the two-dimensional structure of the PlnB membrane domain, the present work investigated PlnB membrane topology by using alkaline phosphatase, β-galactosidase, and β-lactamase gene fusions. We also explored the functionality of the PlnB membrane domain in signal recognition and regulation of kinase activity by construction of hybrid kinase genes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli strains JM109 and CC118, used for plasmid cloning and protein expression, were grown in Luria-Bertani (LB) broth at 37°C with vigorous agitation. Lactobacillus sakei RV2002 and Lb790 were grown in Man-Rogosa-Sharp (MRS) broth (Oxoid) at 30°C without agitation. Where appropriate, ampicillin (100 μg ml−1 for E. coli) or erythromycin (500 μg ml−1 for E. coli and 5 μg ml−1 for L. sakei) was added to the growth medium.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli JM109 | F′ traD36 proA+B+ lacIq Δ(lacZ)M15/Δ(lac-proAB) glnV44 e14−gyrA96 recA1 relA1 endA1 thi hsdR17 | New England Biolabs |
| E. coli CC118 | araD139 Δ(ara leu)7697 ΔlacX74 ΔphoA20 galE galK rspE rpoB argE(Am)recA1 | 27 |
| L. sakei RV2002 | ΔlacLM | 46 |
| L. sakei Lb790 | Wild type, plasmid free; the sppK homologue in this strain contains a mutation in the 5′ end, resulting in a nontranslated transcription product (T. Møretrø, personal communication) | 44 |
| Plasmids | ||
| pCR-topo 2.1 | E. coli cloning vector; Ampr | Invitrogen |
| pMG36e | Shuttle vector containing P32 promoter; Emr | 48 |
| pRMCD28 | E. coli phoA in pWSK29; Ampr | 4 |
| pRMCD70 | E. coli lacZ in pWSK29; Ampr | 4 |
| pELS200 | Shuttle vector; Emr Ampr | L. Axelsson and M. Skaugen (unpublished) |
| pCRAMP | pCR-Blunt II-TOPO with a leaderless blaM gene | This work |
| PJB-G4 | pELS200 with gusA fused to plnA promoter | 8 |
| PJB37-C | pMS37 with plnC fused to P32 promoter | 8 |
| pGH3 | pGEM7Zf(−) containing plnABCD operon | 8 |
| pGC | pELS200 containing gusA reporter unit and plnC response regulator unit | This work |
| pGCB | pGC with gene encoding PlnB | This work |
| pGCHyb1 | pGC with gene encoding H1 | This work |
| pGCHyb2 | pGC with gene encoding H2 | This work |
| pGCHyb3 | pGC with gene encoding H3 | This work |
Construction of phoA and lacZ gene fusions.
Truncated versions of plnB were amplified by PCR with pGH3 as a template. For all constructs, the forward primer contained an XbaI site and was 5′-ATGCTCTAGATTAGAGGTAGTAGCTTGGTTGAAATCAGTATCTTCGAT-3′. For the phoA fusions, the reverse primers contained a HindIII site and were 5′-ATGCAAGCTTTGTTCGTATTACTGAAGATGAAGTT-3′ for N32, 5′-ATGCAAGCTTTCCCCAAAACCAAAATTAGCGAT-3′ for G64, 5′-ATGCAAGCTTTATCGGCATCTAAGTAGATATAC-3′ for D112, 5′-ATGCAAGCTTTCGAGCCTGTGTATCGTCTC-3′ for S153, 5′-ATGCAAGCTTTACTTATATCAAACGAGCCTGTGT-3′ for S157, 5′-ATGCAAGCTTTACCTTGAAAGTTGCTAATAA ATAAC-3′ for G188, 5′-ATGCAAGCTTTTTGAAAGTTGCTAATAAATAAC-3′ for Q187, 5′-ATGCAAGCTTTTCTAATCGTTTCTAATGTTTGCC-3′ for R217, and 5′-ATGCAAGCTTTATAATCATTTAATTGCTTATTCTGC-3′ for Y238. PCR amplification products were digested with XbaI and HindIII and ligated in frame with phoA in the pRMCD28 vector (4). The various reporter plasmids were subsequently transformed into E. coli CC118 by standard procedures (43). The reverse primers used for the lacZ fusions all contained a BamHI site. They were 5′-ATGCGGATCCACGTTCGTATTACTGAAGATGAAGTT-3′ for N32, 5′-ATGCGGATCCACCCCCAAAACCAAAATTAGCGAT-3′ for G64, 5′-ATGCGGATCCACCTGACTAGAAATCAACATTAAGA-3′ for Q90, 5′-ATGCGGATCCACATCGGCATCTAAGTAGATATAC-3′ for D112, 5′-ATGCGGATCCACCGAGCCTGTGTATCGTCTC-3′ for S153, 5′-ATGCGGATCCACACTTATATCAAACGAGCCTGTGT-3′ for S157, 5′-ATGCGGATCCACACCTTGAAAGTTGCTAATAAATAAC-3′ for G188, 5′-ATGCGGATCCACTTGAAAGTTGCTAATAAATAAC-3′ for Q187, 5′-ATGCGGATCCACTCTAATCGTTTCTAATGTTTGCC-3′ for R217, and 5′-ATGCGGATCCACATAATCATTTAATTGCTTATTCTGC-3′ for Y238. PCR products were digested with BamHI and XbaI and ligated in frame with lacZ in the pRMCD70 vector (4). The resulting vectors were subsequently transformed into E. coli CC118. All inserts from PCR were confirmed by DNA sequencing, and correct cloning into final plasmids was verified by restriction analysis. Various lacZ gene fusions from pRMCD70 were digested with KpnI and XbaI and then ligated into pMG36e downstream of the P32 promoter. The resulting vectors were transformed into L. sakei RV2002 by applying the protocol described by Aukrust et al. (1).
Construction of blaM gene fusions.
A blaM gene missing its leader sequence was amplified from pCR-TOPO (Invitrogen), using 5′-ATGCGGATCCTCACCCAGAAACGCTGGTG-3′ as the forward primer (contains a BamHI site at the 5′ end) and 5′-ATGCGGAATTTGACAGTTACCAATGCTTAAT-3′ as the reverse primer. The resulting fragment was inserted into pCR-Blunt II-TOPO (Invitrogen) by blunt-end ligation, resulting in the vector pCRAMP. Next, truncated forms of plnB were amplified by using 5′-ATGCAAGCTTTTAGAGGTAGTAGCTTGGTTGAAATCAGTATCTTCGAT-3′ as the forward primer (contains a 5′-end HindIII restriction site). The reverse primers were the same as for the corresponding lacZ fusion constructs. In addition, four constructs were amplified by using the following reverse primers: 5′-ATGCGGATCCACTTGGCACTTTCCCTTAATAATT-3′ for Q72, 5′-ATGCGGATCCACCAACATTAAGAATACATTTAGATG-3′ for L86, 5′-ATGCGGATCCACCAAAGCTAAAATTACTATCTGAC-3′ for L96, and 5′-ATGCGGATCCACTAAGTAGATATACAGAAATCCTC-3′ for L109. The truncated restriction fragments were digested with HindIII and BamHI and ligated in frame with blaM in pCRAMP. The resulting vectors were transformed into E. coli TOP10 (Invitrogen). The ampicillin resistance of individual cells containing the various plasmids was determined by plating appropriate dilutions on LB agar plates containing from 0 to 60 μg of ampicillin ml−1.
Construction of pGC.
To measure the activities of the various histidine kinases used in this work, the reporter vector pGC was constructed. The vector is composed of a reporter gusA unit, containing the gusA gene fused to the inducible plnA promoter, and a plnC response regulator unit, containing the plnC response regulator gene fused to a constitutive P32 promoter. The two units were obtained from the vectors pJB-G4 and pJB37-C, respectively (8). pJB-G4 and pJB37-C were restricted with suitable restriction enzymes, and the gusA and plnC units were ligated into vector pELS200, giving rise to pGC.
Generation of hybrid histidine kinase genes.
The wild-type kinase gene plnB was fused with the plnA promoter by a two-step PCR (20), using 5′-TCCTAAAAGCGAGGTGATTATTATGGTTGAAATCAGTATCTTCGAT-3′ and 5′-AGGATTTTCGCTCCACTAATAA-3′ (INP2) as internal primers and 5′-CTGCAGAATATATTAATCTAAGTACAGTAC-3′ (EXP1) and 5′-GCGGCCGCTTATTTATCCTCCGTAACAATTAA-3′ (EXP2) as external primers. Wild-type sppK was fused to the same promoter by using 5′-TCCTAAAAGCGAGGTGATTATTATGTTATATACGGATGTATCGGT-3′ and INP2 as internal primers and 5′-GCGGCCGCTTAAGTTTCCTCCGTAATCATC-3′ (EXP3) and EXP2 as external primers. Genomic DNAs from L. plantarum C11 and L. sakei LTH673 were used as templates. The resulting PCR fragments were ligated into the vector pCR-Topo 2.1 (Invitrogen), giving the vectors pCR-PlnB and pCR-SppK, and subsequently sequenced. Next, the hybrid kinase genes hyb1, hyb2, and hyb3 were generated by using these constructs as templates. hyb1 was constructed with 5′-GCTTGGGCTTATTAGTTGGCAAGTGATGCTTTTTTTG-3′ and 5′-GCCAACTAATAAGCCCAAGCATCAAA-3′ as internal primers and EXP1 and EXP2 as external primers. hyb2 was constructed with 5′-GGATGACTTTTTGGCAAACATTAGAAACGATTAGAGT-3′ and 5′-GAATAGCCCTACTGAAAAACCGTTTGTAA-3′ as internal primers and EXP1 and EXP3 as external primers. hyb3 was constructed with 5′-AAGGAGTAACAGCCACAATTCAACTAACTTTACT-3′ and 5′-AATTGTGGCTGTTACTCCTGGAAGATTCCC-3′ as internal primers and EXP1 and EXP2 as external primers. Finally, pCR-PlnB, hyb1, hyb2, and hyb3 were digested with PstI and NotI, and the kinase genes were isolated and subsequently ligated into pGC, giving rise to pGCB, pGCHyb1, pGCHyb2, and pGCHyb3.
Reporter enzyme assays.
Assays of LacZ and PhoA in E. coli were performed as described by Miller (28) and Manoil (27), respectively. Assays of LacZ in L. sakei were performed as described by Stentz et al. (46), with the following modifications; Bacteria were grown in MRS (Oxoid) medium until the optical density at 600 nm reached 0.3. Bacteria from 10 ml of culture were collected by centrifugation and resuspended in 1 ml of Z buffer (100 mM sodium phosphate [pH 7.0], 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol). The bacteria were broken with glass beads in a Fast-Prep bead beater (BIO 101) two times for 20 s each with a 5-min break on ice. Cellular debris was removed by centrifugation, and 200 μl of the supernatant was subsequently mixed with 100 μl of o-nitrophenyl-β-d-galactopyranoside (4 mg/ml) at 28°C. The reaction was stopped by addition of 250 μl of Na2CO3. β-Glucuronidase (GUS) assays were performed as previously described (8), except that cultures were incubated with IP-C11 for 3 h. Assay reaction mixtures were incubated for 30 min at 37°C. The GUS values obtained were used to determine the specific GUS activity (GUS activity/cell density). The ampicillin resistance of individual cells expressing the various PlnB-BlaM fusions was determined by plating appropriate dilutions of exponential-phase cultures onto LB plates containing from 0 to 60 μg of ampicillin ml−1. Following incubation overnight at 37°C, the maximum ampicillin tolerance was estimated by comparing colony numbers on ampicillin-containing plates to colony numbers on control plates. The maximum ampicillin tolerance of a given clone was defined as the ampicillin concentration that reduced colony formation by 50%.
Preparation of protein extracts and immunoblotting techniques.
E. coli cells expressing the various LacZ reporter fusions were grown in LB medium at 37°C to an A600 of 0.4. Cells were harvested from 20 ml of culture and lysed in 500 μl of sodium dodecyl sulfate (SDS) sample buffer at 100°C for 5 min. Proteins were separated by SDS-12% polyacrylamide gel electrophoresis (43). After transfer onto a Hybond-P polyvinylidene difluoride membrane (Amersham), proteins were detected with an anti-LacZ polyclonal antibody (Chemicon) and an anti-rabbit-horseradish peroxidase antibody (Amersham). Blots were developed by enhanced chemiluminescence (ECL; Amersham)
RESULTS
Computer analysis of PlnB membrane topology.
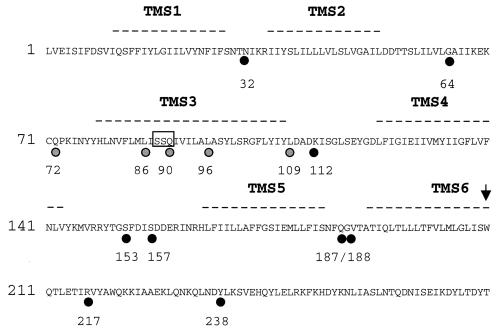
Computer analysis using SOSUI (21), Tmpred (22), and the method of Kyte and Doolittle (24) suggested that the membrane domain of PlnB contains six TMS (Fig. 1). SOSUI predicted an extracytoplasmatic location of the N-terminal tail of TMS1, assigning TMS1 an outside-to-inside (o-i) membrane orientation and TMS2 an inside-to-outside (i-o) membrane orientation. The Tmpred output data showed no significant preference for the orientations of TMS1, TMS2, and TMS5. However, the data showed preferences for an i-o orientation of TMS3 and an o-i orientation of TMS4 and TMS6.
FIG. 1.
Hydropathy analysis of the PlnB membrane domain. Putative TMS identified by SOSUI, Tmpred, and the Kyte-Doolittle algorithm are indicated by dashed lines. Amino acids fused to all three reporter enzymes used in this work (PhoA, LacZ, and BlaM) are indicated by black circles. Gray circles indicate amino acid positions that were investigated only with BlaM. The numbering indicates the positions of the respective residues. An open box indicates the turn-inducing SSQ tripeptide in TMS3. The final C-terminal residue of the transmembrane domain is indicated with an arrow.
Analysis of PlnB membrane topology in E. coli by using plnB-phoA and plnB-lacZ gene fusions.
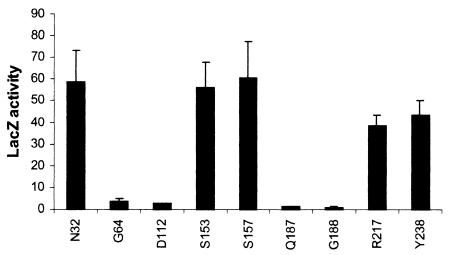
To investigate PlnB membrane topology by genetic approaches, we first used the established technique of alkaline phosphatase (phoA) and β-galactosidase (lacZ) gene fusions (27), in which enzymatically active PlnB-LacZ fusions identify regions in PlnB located in the cytoplasm, whereas active PlnB-PhoA fusions identify regions situated in the periplasm. Based on the computer analysis, we constructed a series of LacZ and PhoA fusions to each of the predicted loops that separate the putative TMS in Fig. 1, with junction sites after N32, G64, D112, S153, S157, Q187, G188, R217, and Y238. The hybrid lacZ and phoA constructs were expressed from a lacI promoter in the low-copy-number vectors pRMCD70 and pRMCD28, respectively (4) (Table 1). A lacZ gene missing the first eight codons was used as a negative control for β-galactosidase activity. Likewise, a phoA gene missing its leader sequence was used as a negative control for alkaline phosphatase activity. As expected, each pair of hybrid proteins demonstrated a contrasting pattern of LacZ and PhoA activity, whereas the negative controls showed no enzymatic activity. As shown in Fig. 2, a reporter fusion to the region connecting the putative TMS1 and TMS2 (N32) resulted in a LacZ+ PhoA− phenotype, indicating a cytoplasmic location of this loop. In accordance with these results, fusions of the reporter proteins to the loop connecting TMS2 and TMS3 (G64) resulted in a LacZ− PhoA+ phenotype, indicating a periplasmic location of this region. These results are in agreement with the SOSUI prediction, assigning TMS1 an o-i orientation and TMS2 an i-o orientation. However, fusions to the loop connecting the putative TMS3 and TMS4 (D112) also showed a LacZ− PhoA+ phenotype. Thus, PhoA fusions at both the N- and C-terminal regions of TMS3 (G64 and D112) exhibited a PhoA+ phenotype. This result led to the hypothesis that either the predicted TMS3 (Fig. 1) does not form a transmembrane segment or, alternatively, this region actually forms two TMS. In accordance with such a model, fusions of the reporter proteins to S153 and S157 resulted in a LacZ+ PhoA− phenotype, suggesting a cytoplasmic location of the loop connecting TMS4 and TMS5, whereas fusions to the loop connecting TMS5 and TMS6 (Q187 and G188) resulted in a LacZ− PhoA+ phenotype, indicating a periplasmic location of these residues. Finally, fusions C terminal of the putative TMS6 (R217 and Y238) displayed a LacZ+ PhoA− phenotype, thus verifying a cytoplasmic location of the C-terminal kinase domain. These results confirmed the presence of TMS4, TMS5, and TMS6 as shown in Fig. 1, suggesting o-i (TMS4), i-o (TMS5), and o-i (TMS6) orientations of the three most C-terminal TMS. To demonstrate that low levels of enzyme activity were due to periplasmic location of the enzyme moiety and not to poor expression of the fusion protein, the production of LacZ fusions was examined by Western blotting. As shown in Fig. 3, low enzymatic activity did not correlate with a low level of expression. For example, the fusions at G64 (LacZ−) and R217 (LacZ+) appeared to be synthesized in equal amounts.
FIG. 2.
Enzymatic activities of PlnB-PhoA and PlnB-LacZ fusions in E. coli CC118. The numbering indicates the terminal residues of PlnB in the different gene fusions. The assays were performed as described in Materials and Methods, and the values represent the means and standard deviations from at least three independent triplicate experiments. LacZ activities are indicated in Miller units (28), whereas results for the PhoA assays are indicated in alkaline phosphatase units (27).
FIG. 3.
Western immunoblots of E. coli cultures expressing various LacZ reporter fusions. Total proteins from cells harboring the PlnB-LacZ fusions were separated by SDS-12% polyacrylamide gel electrophoresis and detected by Western blotting with an anti-LacZ antibody (Chemicon) and an anti-rabbit antibody conjugated with horseradish peroxidase (Amersham). The blots were developed by ECL (Amersham). The PlnB-LacZ fusions shown are to amino acids 32 (lane 1), 64 (lane 2), 90 (lane 3), 112 (lane 4), 153 (lane 5), and 217 (lane 6).
Analysis of PlnB membrane topology in E. coli by using plnB-β-lactamase gene fusions.
C-terminal deletion fusions to β-lactamase (BlaM) have previously been used successfully to resolve the topology of prokaryotic membrane proteins (2). When grown as single colonies, cells expressing fusion proteins with the β-lactamase moiety fused to a cytoplasmic protein domain show low tolerance to the presence of ampicillin, because cytoplasmic β-lactamase cannot intervene between the antibiotic and its periplasmic targets. In comparison, cells expressing fusion proteins in which β-lactamase is fused to a periplasmic protein domain will be less sensitive to ampicillin, since the periplasmic location of the β-lactamase enzyme protects the cells against the antibiotic. To further characterize the membrane topology of PlnB, 14 plnB-blaM gene fusions were constructed. This was facilitated by amplification of truncated fragments of plnB by PCR and subsequent ligation of these fragments in frame with a leaderless blaM gene in pCRAMP (for details, see Materials and Methods). The resulting constructs were subsequently transformed into E. coli Top 10 cells (Invitrogen). To decide the cytoplasmic or periplasmic location of the PlnB-β-lactamase fusion junction in each construct, cells harboring the constructs were grown as single colonies on a series of LB agar plates with various concentrations of ampicillin. The pattern of ampicillin resistance of the fusions agreed with the results obtained with the PhoA-LacZ reporter system. As shown in Fig. 4, β-lactamase fusions to N32, S153, S157, R217, and Y238 displayed sensitivity to low concentrations of ampicillin, suggesting a cytoplasmic location of the fusion junctions in these constructs. In comparison, fusions to G64, Q72, D112, Q187, and G188 displayed an increased resistance to ampicillin, indicating a periplasmic location of the fusion junctions. As shown in Fig. 1, the N- and C-terminal parts of the putative TMS3 are separated by a polar tripeptide sequence (SSQ), which could contribute to a turn structure (42). To determine the topology of the TMS3 region, we next constructed β-lactamase fusions to L86, Q90, L96, and L109. Fusions N terminal of the SSQ tripeptide (L86 and Q90) showed ampicillin sensitivity comparable to that of the fusion at S153 (Fig. 4), indicating that the constructs have the β-lactamase moiety fused to a protein segment pointing toward the cytoplasmic side of the membrane. In contrast, fusions C terminal of the SSQ tripeptide region (L96 and L109) conferred significant ampicillin resistance. As shown in Fig. 4, the level of ampicillin sensitivity in cells expressing these proteins decreased with increased distance between the SSQ tripeptide and the fusion junction of the proteins. The data indicated that the C-terminal region of TMS3 in Fig. 1, in contrast to the N-terminal region, points toward the periplasm, suggesting that the SSQ part of TMS3 induces a turn in the polypeptide chain. To determine the location of this turn, we next constructed PlnB-LacZ and PlnB-PhoA fusions with junctions at Q90. The PlnB-LacZ fusion displayed significant enzymatic activity, whereas no activity was observed with the PhoA fusion (Fig. 2), suggesting a cytoplasmic location of the SSQ turn. Similar to the case for the PlnB-LacZ fusions, the enzymatic activities of the various β-lactamase fusions did not appear to correlate with the amount of expression (data not shown). Taken together, the gene fusion data led to the topological model illustrated in Fig. 4, in which the membrane domain of PlnB contains seven TMS.
FIG. 4.
Model of PlnB membrane topology based on PhoA, LacZ, and BlaM fusions. Gray circles indicate residues fused to BlaM, while the numbering indicates the positions of the respective residues. Numbers in brackets correspond to the maximum ampicillin tolerance (in micrograms per milliliter) conferred by the respective fusions. Positively charged residues that could act as potential topological determinants are indicated by +. Note that the residues corresponding to the cytoplasmic kinase domain are not shown.
Expression of LacZ fusions in Lactobacillus.
To confirm that the topological data obtained for the PlnB membrane topology in E. coli also reflect the membrane conformation that PlnB adopts in Lactobacillus, a selection of lacZ fusions were digested from pRMCD70. These fragments were ligated downstream of the constitutive P32 promoter in the broad-host-range vector pMG36e. The resulting plasmids were subsequently transformed into L. sakei RV2002, which carries a deletion of the lacLM operon (46). As shown in Fig. 5, the fusion constructs displayed a pattern of LacZ activity in L. sakei similar to that observed in E. coli. The corresponding PhoA fusions were not tested in L. sakei, since folding of PhoA requires the formation of intramolecular disulfide bonds by a periplasmic E. coli enzyme system (45). However, the L. sakei LacZ fusion data present convincing evidence that the topological model of PlnB derived from gene fusion experiments in E. coli is valid in Lactobacillus.
FIG. 5.
Enzymatic activities of PlnB-LacZ fusions in L. sakei RV2002. The numbering indicates the terminal residues of PlnB in the different gene fusions. The assays were performed as described in Materials and Methods, and the values represent the means and standard deviations from at least three independent triplicate experiments. LacZ activities are indicated in Miller units (28).
The transmembrane domain of PlnB functions as a receptor for the peptide pheromone.
Having established a topological model for the PlnB membrane domain, we next investigated how this domain influences the activity of the cytoplasmic kinase domain. For this purpose, a reporter system to measure PlnB kinase activity in vivo was constructed (Fig. 6, upper panel). First, a gusA reporter gene was fused to a plnA promoter fragment. Next, the plnC response regulator gene was fused to a constitutive lactococcal P32 promoter. These two units were ligated into the vector pELS200, giving rise to pGC. Finally, the gene encoding plnB was fused to a plnA promoter fragment and ligated into pGC (giving rise to pGCB). pGC and pGCB were subsequently transformed into the heterologous host L. sakei Lb790. In cells harboring pGCB, transcription of the gusA reporter unit is regulated by PlnC, which upon phosphorylation binds to and activates the plnA promoter. Consequently, the level of expression from the gusA reporter monitors the rate at which PlnB phosphorylates PlnC. As shown in Fig. 6 (lower panel), no increase of GUS activity was observed in cells containing pGC upon addition of the PlnB peptide inducer pheromone IP-C11 (NH2-KSSAYSLQMGATAIKQVKKLFKKWGW-COOH). Similarly, no increase in GUS activity was observed when cells harboring pGC or pGCB were induced with IP-673 (NH2-MAGNSSNFIHKIKQIKTHR-COOH), a peptide pheromone corresponding to the PlnB homologue SppK from L. sakei LTH673 (11). In contrast, cells containing pGCB showed an approximately threefold increase of GUS activity when induced with IP-C11, indicating an increased level of phosphorylated PlnC in these cells. Next, a hybrid kinase gene (hybI) composed of the membrane domain from sppK and the kinase domain from plnB was constructed (Fig. 6, upper panel). hyb1 was ligated into pGC, giving rise to pGCHyb1. When IP-673 was added to the growth medium of cells harboring this construct, a threefold increase in GUS activity was observed. No increase of GUS activity was observed upon addition of IP-C11. In contrast, when a hybrid gene (hyb2) containing the transmembrane domain from plnB and the kinase domain from sppK was ligated into pGC (giving rise to pGCHyb2), increased GUS activity was observed only when cells were induced with IP-C11 (Fig. 6, lower panel). It has previously been shown that the SppR response regulator is able to bind to the plnA promoter (41) It is therefore unclear whether the Hyb2 kinase domain phosphorylates the plasmid-encoded PlnC response regulator or the chromosomally encoded SppR regulator present in L. sakei Lb790. Nevertheless, the results conclusively show that the IP receptors of PlnB and SppK are located in the transmembrane portion of the proteins. The results also suggest that the mode of signal transduction from the ligand-bound IP receptor to the kinase domain proceeds through similar mechanisms in PlnB and SppK. To further investigate the molecular basis for the specificity of peptide pheromone recognition in SppK and PlnB, one additional hybrid kinase gene (hyb3) was constructed. This hybrid contained a PlnB kinase domain and an SppK membrane domain in which the most C-terminal extracytoplasmatic loop and TMS were exchanged with the corresponding sequence from PlnB (Fig. 6, upper panel). As shown in Fig. 6 (lower panel), this hybrid responded to addition of IP-673 but not to addition of IP-C11. This result indicates that determinants for IP interaction in SppK and PlnB are located N terminal of the final extracytoplasmatic loop of the transmembrane domain. Interestingly, compared to Hyb1 and Hyb2, Hyb3 showed an elevated level of basal kinase activity in the absence of peptide pheromone (discussed below).
FIG. 6.
Kinase activities of hybrid kinases. (Upper panel) Schematic presentation of the reporter system used to measure kinase activity in vivo. The system contains a reporter unit (gusA) fused to the plnA promoter (arrow) and a response regulator unit (plnC) fused to the constitutive P32 lactococcal promoter (arrow). Transcription from the plnA promoter in the reporter unit is regulated by the phosphorylation status of PlnC. plnB, hyb1, hyb2, and hyb3 were ligated into pGC, giving rise to the plasmids pGCB and pGCHyb1 to -3, respectively. The sites of PlnB-SppK fusion junctions were W210 in Hyb1 and Hyb2 and F176 in Hyb3 (indicated above each hybrid). Both W210 and F176 are conserved in PlnB and SppK. White segments in hybrid genes indicate sequences corresponding to SppK, while gray segments correspond to sequences derived from PlnB. All vectors were subsequently transformed into L. sakei Lb790. (Lower panel) GUS activities (means and standard deviations) in Lb790 clones harboring pGC, pGCB, pGCHyb1, pGCHyb2, and pGCHyb3. Cells were induced at an A600 of 0.15 with 500 ng of IP-C11 or IP-673 ml−1 and incubated at 30°C for 3 h. Addition of IP at a higher concentration did not result in a further increase of GUS activity. Specific GUS activity (GA) was calculated as A405/A600.
DISCUSSION
Peptide pheromone-activated histidine kinases from the HPK10 subfamily control several important biological processes in gram-positive bacteria, including competence for genetic transformation, bacteriocin synthesis, and virulence. Such regulatory systems, including the com, pln, and agr regulons, have been extensively studied with respect to the events following phosphorylation of the response regulators (12, 18, 23, 35). However, less is known about the interaction between the various peptide pheromones and their cognate kinase receptors. In this work we have characterized the membrane topology of the PlnB receptor domain by using reporter gene fusions. Six hydrophobic segments were initially identified by computer analysis of PlnB (Fig. 1), whereas the data obtained with alkaline phosphatase, β-galactosidase, and β-lactamase fusions in both E. coli and L. sakei suggested a topological model in which the PlnB membrane domain contains seven TMS. The reporter fusion data agreed with the computer approximations in that they identified the predicted TMS1 and -2 and TMS4 to -6 (Fig. 1) as authentic transmembrane segments. However, the experimental data suggested that the predicted TMS3 in Fig. 1 forms two TMS (Fig. 4), with a cytoplasmic turn being induced by the SSQ tripeptide located in the middle of this region. In support of the topological model illustrated in Fig. 4, it has recently been demonstrated that short repeats of polar residues can induce tight turns between closely spaced TMS, a trait not recognized by current prediction algorithms (29, 42). The overall topological model of PlnB is in agreement with the positive-inside rule (49, 50), containing the majority of positively charged residues on the cytoplasmic side of the membrane. Conspicuously, the most N-terminal extracytoplasmatic loop contains three lysine residues. However, site-directed mutagenesis in which Lys68, Lys70, or Lys74 was replaced with Ala did not affect wild-type PlnB function, indicating that the positive charges in this region are not important for determining PlnB topology (data not shown). Since it is very difficult to determine the exact boundaries of a TMS experimentally, the TMS endpoints in Fig. 4 are based upon the computer predictions for PlnB and should be considered putative. Members of the HPK10 subfamily have very similar hydropathy profiles, and it has previously been speculated that these proteins contain membrane domains with related or identical two-dimensional topologies. Interestingly, computer-aided predictions for ComD using a sequence alignment of ComD proteins from six species of Streptococcus resulted in a topological model more or less identical to the model presented for PlnB in Fig. 4 (18). Furthermore, a previous study indicated that the C-terminal part of the AgrC membrane domain also adopts a conformation very similar to that of PlnB (25). Unfortunately, the topology of the 100 to 120 amino acids at the N-terminal end of AgrC could not be resolved. However, similar to the case for PlnB and ComD, this region in AgrC is predicted to contain three or four TMS. If the HPK10 kinases adopt similar topologies in their membrane domain, it would be reasonable to assume that the signal transduction from the IP receptor to the catalytic domain proceeds through a conserved mechanism. Our results with hybrid kinase proteins suggest that this could be the case. Although they are from bacteria of the same genus (Lactobacillus), PlnB and SppK share only 22% sequence identity in their transmembrane domains, while the corresponding peptide pheromones (IP-C11 and IP-673, respectively) show little or no sequence identity at all (23). Still, as observed with Hyb1, the membrane domain of SppK efficiently regulated the enzymatic activity of the PlnB kinase domain. The SppK membrane domain not only was able to repress the activity of the PlnB kinase domain in the absence of IP but also efficiently stimulated kinase activity upon addition of IP-673. Further, when a hybrid kinase containing the transmembrane portion of PlnB and the kinase domain of SppK (Hyb2) was constructed, kinase activity could be induced by addition of IP-C11. These results clearly demonstrate that all determinants for inducer specificity are contained within the N-terminal transmembrane portion of the proteins. Perhaps more interestingly, the results also indicate that the conformational change that the SppK receptor domain undergoes upon binding of IP-673 is transduced to the kinase domain in a manner that mimics signaling in wild-type PlnB. These observations reinforce the hypothesis that the mode of signal transduction in different HPK10 kinases is conserved, and they thus point toward a common structural and functional organization of the transmembrane receptor domains within this subfamily. Still, one must be aware that the concept of a common structural organization in the HPK10 subfamily cannot be fully proven until additional structural data are produced for other family members.
Little information about the nature of the conformational change that the receptor domain of HPK10 kinases undergoes upon binding of the peptide pheromone has been published. Previous studies of N-terminal deletion variants of AgrC fused to the E. coli maltose-binding protein proposed that the final extracytoplasmatic loop and TMS of the protein prevent spontaneous activity of the kinase domain in the absence of peptide pheromone (25). When this region of SppK was exchanged with the corresponding residues from PlnB (Hyb3), a significant increase in the basal level of kinase activity was observed. Similar to the observations for AgrC, this could indicate that the positioning of the most C-terminal TMS in SppK and PlnB relative to the rest of the protein is critical for determining the activity of the cytoplasmic kinase domain. As shown in Fig. 6 (lower panel), Hyb3 could be activated, although weakly, by addition of the SppK-specific IP-673 to the growth medium. The weak increase in activity upon addition of IP-673 is perhaps not surprising, since the kinase domain of Hyb3 is almost fully activated in the absence of IP. In contrast, no significant response was seen upon addition of IP-C11. These results indicate that determinants for SppK and PlnB IP specificity are located N terminal of the final extracytoplasmatic loop in Fig. 4. Interestingly, sequence alignments of ComD kinases from Streptococcus pneumoniae Rx (ComD-1) (36) and S. pneumoniae A66 (ComD-2) show that the two proteins differ in only 12 amino acid positions within the transmembrane domain (data not shown). Despite the high level of sequence identity in the receptor domains of these kinases, the corresponding peptide pheromones do not cross-activate between ComD-1 and ComD-2 (37). According to the above-mentioned topological prediction for ComD, seven of the substituted residues cluster to the C-terminal region of the second most N-terminal TMS. It is therefore tempting to speculate that this region is important for deciding the IP specificity of the ComD receptors. In line with such a hypothesis, point mutations in this region of PlnB also seem to drastically alter the ability of IP-C11 to activate PlnB (O. Johnsborg and I. F. Nes, unpublished data). We are currently undertaking experiments to further assess the putative involvement of this region in PlnB IP interaction.
Acknowledgments
We thank G. von Heijne, L. S. Håvarstein, M. Skaugen, and D. A. Brede for helpful discussions and L. Godager for technical assistance.
This work was supported by grants from the Research Council of Norway.
REFERENCES
- 1.Aukrust, T. W., M. B. Bruberg, and I. F. Nes. 1995. Transformation of Lactobacillus by electroporation. Methods Mol. Biol. 47:201-208. [DOI] [PubMed] [Google Scholar]
- 2.Broome-Smith, J. K., M. Tadayyon, and Y. Zhang. 1990. Beta-lactamase as a probe of membrane protein assembly and protein export. Mol. Microbiol. 4:1637-1644. [DOI] [PubMed] [Google Scholar]
- 3.Bruberg, M. B., I. F. Nes, and V. G. H. Eijsink. 1997. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol. Microbiol. 26:347-360. [DOI] [PubMed] [Google Scholar]
- 4.Daniels, C., C. Windurampulle, and R. Morona. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol. Microbiol. 28:1211-1222. [DOI] [PubMed] [Google Scholar]
- 5.Diep, D. B., L. S. Håvarstein, J. N. Meyer, and I. F. Nes. 1994. The gene encoding plantaricin A, a bacteriocin from Lactobacillus plantarum C11, is located on the same transcription unit as an agr-like regulatory system. Mol. Microbiol. 60:160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diep, D. B., L. S. Håvarstein, and I. F. Nes. 1995. A bacteriocin-like peptide induces bacteriocin synthesis in Lactobacillus plantarum C11. Mol. Microbiol. 18:631-639. [DOI] [PubMed] [Google Scholar]
- 7.Diep, D. B., L. S. Håvarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178:4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diep, D. B., O. Johnsborg, P. A. Risøen, and I. F. Nes. 2001. Evidence for dual functionality of the operon plnABCD in the regulation of bacteriocin production in Lactobacillus plantarum. Mol. Microbiol. 41:633-644. [DOI] [PubMed] [Google Scholar]
- 9.Diep, D. B., R. Myhre, O. Johnsborg, Å. Åkra, and I. F Nes. 2003. Inducible bacteriocin production in Lactobacillus is regulated by differential expression of the pln operons and by two antagonizing response regulators, the activity of which is enhanced upon phosphorylation. Mol. Microbiol. 47:483-494. [DOI] [PubMed] [Google Scholar]
- 10.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in Gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 11.Eijsink, V. G. H., M. B. Bruberg, P. H. Middelhoven, and I. F. Nes. 1997. Induction of bacteriocin production in Lactobacillus sake by a secreted peptide. J. Bacteriol. 178:2232-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eijsink, G. H. V., L. Axelsson, D. B. Diep, L. S. Håvarstein, H. Holo, and I. F. Nes. 2002. Production of class II bacteriocins by lactic acid bacteria; and example of biological warfare and communication. Antonie Leeuwenhoek 81:639-654. [DOI] [PubMed] [Google Scholar]
- 13.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 14.Guangyong, J., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Håvarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 16.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 17.Håvarstein, L. S., and D. A Morrison. 1999. Quorum sensing and peptide-pheromones in streptococcal competence for genetic transformation, p. 9-26. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 18.Håvarstein, L. S. 2002. Intercellular communication in Gram-positive bacteria depends on peptide pheromones and their histidine kinase receptors, p. 341-363. In M. Inouye (ed.), Histidine kinases in signal transduction. Academic Press, New York, N.Y.
- 19.Hauge, H. H., D. Mantzilas, G. N. Moll, W. N. Konings, A. J. Driessen, V. G. Eijsink, and J. Nissen-Meyer. 1998. Plantaricin A is an amphiphilic alpha-helical bacteriocin-like pheromone which exerts antimicrobial and pheromone activities through different mechanisms. Biochemistry 37:16026-16032. [DOI] [PubMed] [Google Scholar]
- 20.Higuchi, R. 1990. PCR protocols. A guide to methods and applications, p. 177-183. Academic Press, San Diego, Calif.
- 21.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann, K. W., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 23.Kleerebezem, M., and L. E. Quadri. 2001. Peptide pheromone-dependent regulation of antimicrobial peptide production in Gram-positive bacteria: a case of multicellular behavior. Peptides 22:1579-1596. [DOI] [PubMed] [Google Scholar]
- 24.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 25.Lina, G., S. Jarraud, G. Ji, T. Greenland, A. Pedraza, J. Etienne, R. P. Novick, and F. Vandenesch. 1998. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 12:655-662. [DOI] [PubMed] [Google Scholar]
- 26.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761-777. [DOI] [PubMed] [Google Scholar]
- 27.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and β-galactosidase gene fusions. Methods Cell. Biol. 34:61-75. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 29.Monne, M., M. Hermansson, and G. von Heijne. 1999. A turn propensity scale for transmembrane helices. J. Mol. Biol. 288:141-145. [DOI] [PubMed] [Google Scholar]
- 30.Nes, I. F., D. B. Diep, L. S. Havarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek. 70:113-128. [DOI] [PubMed] [Google Scholar]
- 31.Nes, I. F., and G. H. V Eijsink. 1999. Regulation of group II peptide bacteriocin synthesis by quorum-sensing mechanisms, p. 175-192. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 32.Nilsen, T., I. F. Nes, and H. Holo. 1998. An exported inducer peptide regulates bacteriocin production in Enterococcus faecium CTC492. J. Bacteriol. 180:1848-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ninfa, A. J. 1991. Protein phosphorylation and the regulation of cellular processes by the homologous two-component regulatory systems of bacteria, p. 39-72. In J. K. Setlow (ed.), Genetic engineering, vol. 13. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 34.Novick, R. P., S. J. Projan, J. Kornblum, H. F. Ross, G. Ji, B. Kreiswirth, et al. 1995. The agr P2 operon, an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446-458. [DOI] [PubMed] [Google Scholar]
- 35.Novick, R. P., and T. W. Muir. 1999. Virulence gene regulation by peptides in staphylococci and other Gram-positive bacteria. Curr. Opin. Microbiol. 2:40-45. [DOI] [PubMed] [Google Scholar]
- 36.Pestova, E. V., L. S. Håvarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 37.Pozzi, G., L. Masala, F. Ianelli, R. Manganelli, L. S. Håvarstein, L. Piccoli, D. Simon, and D. A. Morrison. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quadri, L. E., M. Kleerebezem, O. P. Kuipers, W. M. de Vos, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1997. Characterization of a locus from Carnobacterium piscicola LV17B involved in bacteriocin production and immunity: evidence for a global inducer-mediated transcriptional regulation. J. Bacteriol. 179:6163-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Risøen, P. A., L. S. Håvarstein, D. B. Diep, and I. F. Nes. 1998. Identification of the DNA-binding sites for two response regulators involved in control of bacteriocin synthesis in Lactobacillus plantarum C11. Mol. Gen. Genet. 259:224-232. [DOI] [PubMed] [Google Scholar]
- 40.Risøen, P. A., M. B. Brurberg, V. G. H. Eijsink, and I. F. Nes. 2000. Functional analysis of promoters involved in quorum sensing-based regulation of bacteriocin production in Lactobacillus. Mol. Microbiol. 37:619-628. [DOI] [PubMed] [Google Scholar]
- 41.Risøen, P. A., O. Johnsborg, D. B. Diep, L. Hamoen, G. Venema, and I. F. Nes. 2001. Regulation of bacteriocin production in Lactobacillus plantarum depends on a conserved promoter arrangement with consensus binding sequence. Mol. Gen. Genet. 265:198-206. [DOI] [PubMed] [Google Scholar]
- 42.Saaf, A., M. Hermansson, and G. von Heijne. 2000. Formation of cytoplasmic turns between two closely spaced transmembrane helices during membrane protein integration into the ER membrane. J. Mol. Biol. 301:191-197. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Schillinger, U., and F. K. Lucke. 1989. Antibacterial activity of Lactobacillus sake isolated from meat. Appl. Environ. Microbiol. 55:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sone, M., S. Kishigami, T. Yoshihisa, and K. Ito. 1997. Roles of disulfide bonds in bacterial alkaline phosphatase. J. Biol. Chem. 272:6174-6178. [DOI] [PubMed] [Google Scholar]
- 46.Stentz, R., C. Loizel, C. Malleret, and M. Zagorec. 2000. Development of genetic tools fro Lactobacillus sakei: disruption of the β-galactosidase gene and use of lacZ as a reporter gene to study regulation of the putative copper ATPase, AtkB. Appl. Environ. Microbiol. 66:4272-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stock, J. B., M. G. Surette, M. Levit, and P. Park. 1995. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis, p. 25-51. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 48.Van de Guchte, M., J. M. van der Vossen, J. Kok, and G. Venema. 1989. Construction of a lactococcal expression vector. Expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 53:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Heijne, G. 1989. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature 341:456-458. [DOI] [PubMed] [Google Scholar]
- 50.von Heijne, G. 1992. Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]