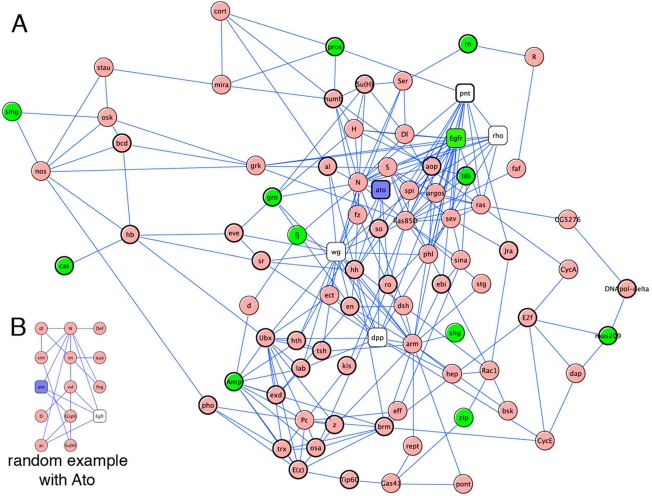
Figure 6. Protein-protein association subnetworks.
The subgraphs are extracted from STRING connections with confidence score above 0.842, and aim to connect as many seed genes as possible. Seed genes may be connected through maximally two edges from either side. (A) The 16 positive ‘known’ genes are used as seed genes to validate their potential relationship. The resulting network is significantly larger and more interconnected than expected by chance and recovers Ato itself as member of the sub-network. (B) Example of a subgraph generated from a random selection of seed genes that does recover Ato. Only 29 out of 1000 random networks recover Ato by chance. These networks are significantly smaller than the network formed by the 16 genes from the screen. Green nodes are positive genes from our screen, while all other nodes are drawn from STRING interaction data (square nodes were part of the training set).