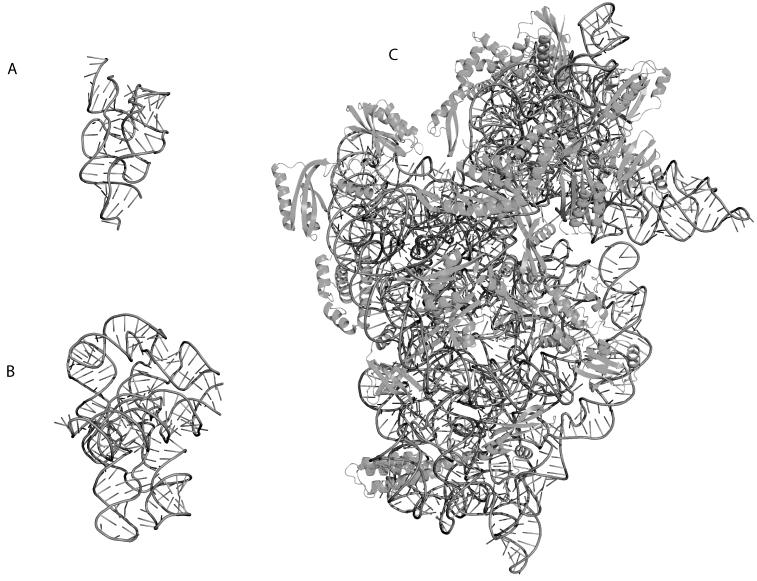
Figure 1.
Three-dimensional crystal structure cartoon representations of functionally important RNAs rendered in PYMOL (http://pymol.sourceforge.net/). The RNA backbone is indicated as a dark grey tube, while the sticks indicate the orientation of the bases relative to the backbone. A) Structure of a Riboswitch (PDB ID 2H0M) bound to Thiamine. This molecule senses Thiamine and undergoes a conformational change when the ligand is present, thereby down-regulating Thiamine synthesis. B) Structure of the Twort group I intron (PDB ID 1Y0Q). This catalytic RNA auto-splices itself thereby processing the mRNA for translation. C) Structure of the T. thermophilus 30S Ribosomal Subunit (PDB ID 1J5E). The proteins are indicated in dark grey and drawn using cartoon representations. This rRNA forms a molecular machine with the 50S subunit that captures the mRNA and translates it into the functional protein.