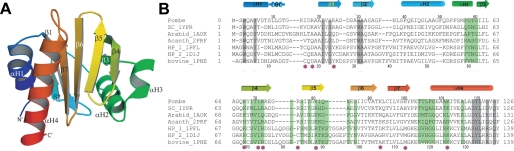
FIGURE 3.
Structure of fission yeast profilin. A, ribbon diagram of S. pombe profilin showing the secondary structure with the four helices labeled αH1–αH4 and seven β-strands labeled β1–β7. B, comparison of profilin sequences aligned based on crystal structures with α-helices noted as cylinders, β-strands noted as arrows, and residues implicated by mutagenesis in phosphatidylinositol 4,5-bisphosphate binding noted with purple circles. SC, S. cerevisiae profilin (PDB 1YPR); Arabid, Arabidopsis thaliana profilin (PDB 1AOK); Acanth, A. castellanii profilin (PDB 2PRF); HP_1, Hs profilin-I (PDB 1PFL); HP_2, Hs profilin-II (1DIJ); and bovine profilin (PDB 1PNE). Structural alignments were done with the O program. Highlights show residues that interact with actin (green) or polyproline (gray) in crystal structures. Residue numbers for S. pombe profilin are shown above the sequences, whereas those of bovine profilin are shown below the sequence alignment.