Abstract
DEAD-box proteins are ubiquitous in RNA metabolism and use ATP to mediate RNA conformational changes. These proteins have been suggested to use a fundamentally different mechanism from the related DNA and RNA helicases, generating local strand separation while remaining tethered through additional interactions with structured RNAs and RNA-protein (RNP) complexes. Here, we provide a critical test of this model by measuring the number of ATP molecules hydrolyzed by DEAD-box proteins as they separate short RNA helices characteristic of structured RNAs (6–11 bp). We show that the DEAD-box protein CYT-19 can achieve complete strand separation using a single ATP, and that 2 related proteins, Mss116p and Ded1p, display similar behavior. Under some conditions, considerably <1 ATP is hydrolyzed per separation event, even though strand separation is strongly dependent on ATP and is not supported by the nucleotide analog AMP-PNP. Thus, ATP strongly enhances strand separation activity even without being hydrolyzed, most likely by eliciting or stabilizing a protein conformation that promotes strand separation, and AMP-PNP does not mimic ATP in this regard. Together, our results show that DEAD-box proteins can disrupt short duplexes by using a single cycle of ATP-dependent conformational changes, strongly supporting and extending models in which DEAD-box proteins perform local rearrangements while remaining tethered to their target RNAs or RNP complexes. This mechanism may underlie the functions of DEAD-box proteins by allowing them to generate local rearrangements without disrupting the global structures of their targets.
Keywords: CYT-19, group I intron, RNA chaperone, RNA folding, RNA helicase
Structured RNAs and RNA-protein complexes (RNPs) mediate a host of essential cellular processes, including processing of messenger RNAs and their translation into protein. In addition to folding into defined structures, many of these RNAs and RNPs undergo extensive conformational changes during their functions. Both their initial folding and conformational changes typically require DEAD-box proteins, which use ATP to promote RNA structural transitions.
DEAD-box proteins are members of helicase superfamily-2 (SF2) and are related to the ATP-dependent RNA and DNA helicases that function in replication and other aspects of nucleic acid metabolism (1). However, rather than unwinding long, continuous duplexes, many DEAD-box proteins manipulate highly structured RNAs and RNPs by facilitating rearrangements that can include local disruptions of secondary structure, tertiary structure, and RNA-protein interactions.
Consistent with their distinct functions, recent in vitro studies have strongly suggested that DEAD-box proteins operate on structured RNAs by a mechanism that is fundamentally different from processive helicases. The Neurospora crassa CYT-19 protein is required for proper folding of several mitochondrial group I introns in Neurospora crassa (2). It also assists folding of diverse group I and group II introns in vitro or when expressed in yeast (3–5), indicating that it acts as a general RNA chaperone.
To gain insight into the mechanism of its activity, we took advantage of the observation that CYT-19 can use its nonspecific chaperone activity to efficiently separate the 6-base pair helix termed P1, formed between group I introns and their 5′-exon-intron junction, and that this unwinding efficiency (kcat/KM) is enhanced by 2 orders of magnitude when the duplex is covalently linked to the ribozyme compared with the same duplex in solution (4). Then, using simple constructs based on group I intron structure, we found that the activity was also enhanced by simple extensions to the helix. Interestingly, a single-stranded extension gave a smaller enhancement than a double-stranded flanking region, and both gave smaller enhancements than the highly structured intact group I intron.
Whereas conventional DNA and RNA helicases commonly require a 5′- or 3′-single-stranded region, which serves as a starting point for translocation into and through the duplex, results above suggested that the increased activity arose instead from an additional and distinct interaction of CYT-19 with the RNA. Further, the enhancement under subsaturating conditions (kcat/KM) suggested that this interaction is maintained in the transition state for strand separation. Additional work showed that the enhancement is nearly eliminated by deletion of 49 aa from the highly basic C terminus of CYT-19, whereas strand separation of a duplex that lacks an extension is essentially unaffected, most simply suggesting that this additional interaction is mediated by the C-terminal region (6).
Together, these findings led to a model in which interactions with adjacent RNA structure tether DEAD-box proteins in proximity to exposed helices or perhaps other elements of RNA structure, where binding of the core domain and ATP-dependent conformational changes give strand separation (4, 7, 8). Although early studies demonstrating that DEAD-box proteins can readily separate duplexes of ≈1 helical turn or less but are essentially inactive for duplexes of 2 or more turns (9, 10) indicated a lack of processivity, the tethering model suggests a more radical difference in mechanism from processive helicases. This is because continuous formation of a tethering interaction during duplex unwinding would most simply suggest the absence of any translocation during the unwinding process.
Strong independent support for essential features of this model has come from studies in the Jankowsky laboratory, using an elegant set of model duplex substrates. First, they demonstrated conclusively that a flanking sequence can enhance activity for DEAD-box proteins without serving as a starting point for translocation by showing that a single-stranded segment can still give activation if it is not linked to the target duplex but is instead bound through biotin-mediated interactions with the protein streptavidin (7). Second, they showed that even model duplexes with an RNA segment flanked on both sides by DNA can be efficiently separated by DEAD-box proteins, whereas the same proteins are not active on fully DNA substrates, indicating that strand separation can be initiated internally, without translocation (11). The main features of the model are also indicated for the Escherichia coli DEAD-box protein DbpA by studies from Uhlenbeck and colleagues, with the important difference that this protein uses an ancillary domain to recognize a particular structure within the large subunit ribosomal RNA rather than interacting with structured RNA more generally (12–15).
In the current work, we have tested and extended this model by measuring the number of ATP molecules used by CYT-19 and other DEAD-box proteins as they separate RNA duplexes. In the most extreme form of the model, with strand separation accomplished in the presence of a continuously formed tethering interaction, it would be possible that the complete reaction would be accomplished in a single cycle of ATP-dependent conformational changes and would therefore give hydrolysis of only 1 ATP. Indeed, we obtain this result for duplexes of 6–11 bp, characteristic of helices present in structured RNAs. Further, under some conditions, a significant fraction of strand-separation events occur in the absence of any ATP hydrolysis. Nevertheless, these events are dependent on ATP, indicating that bound ATP favors a protein conformation that promotes local strand separation even before ATP hydrolysis.
Results
In designing a duplex substrate for these studies, we took advantage of our previous results that CYT-19 can efficiently separate the P1 duplex of the Tetrahymena group I ribozyme, leading to dissociation of the oligonucleotide substrate, and the separation is much more efficient when P1 is covalently linked to the intron or to another RNA helix (4). This increased activity allows robust experimental signals for strand separation and duplex-dependent ATPase activity (ref. 4 and results herein). However, attachment of the intron or even a second RNA duplex would inextricably complicate the analysis because the additional RNA would be expected to interact with CYT-19 and stimulate its ATPase activity, and it would not be possible to determine how much of the total ATPase activity arose from separation of the P1 duplex.
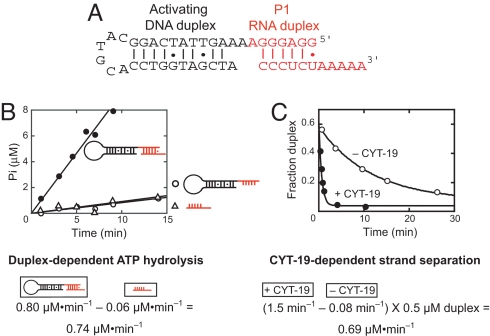
We therefore generated a construct in which the P1 duplex is formed from an RNA oligonucleotide (CCCUCUA5) and a hybrid RNA/DNA oligonucleotide, resulting in P1 being flanked by a DNA duplex (Fig. 1A). This dsDNA extension gave the same activation of strand separation activity as an equivalent RNA extension and, as expected from earlier work (16, 17), an oligonucleotide containing only the DNA portion did not stimulate ATPase activity, implying that it is not actively unwound by the helicase core (Fig. S1).
Fig. 1.
ATP hydrolysis and RNA strand separation by CYT-19. (A) Structure of the duplex substrate, derived from the P1 duplex of the Tetrahymena group I intron. RNA nucleotides are red and DNA nucleotides are black. The DNA portion has the equivalent sequence of the P2 helix, which is adjacent to P1 in the natural intron. (B) P1 duplex-dependent ATP hydrolysis by CYT-19. ATP hydrolysis was measured in the presence of the P1 duplex by including both oligonucleotides (●), or with the same concentrations of the RNA/DNA oligonucleotide (0.5 μM, ○) or the excess CCCUCUA5 (1 μM, triangles) alone. (C) Strand separation of the P1 duplex construct in the presence (filled circles) or absence (open circles) of CYT-19. Excess CCCUCUA5 (5 μM) was present to prevent reannealing of the 32P-labeled CCCUCUA5. An equivalent reaction with 1 μM CCCUCUA5 gave the same rate constant within error (Fig. S3), but the higher concentration allowed more precision by increasing the extent of displacement of the labeled CCCUCUA5. Experimental conditions for B and C were 25 °C, pH 7.0, 10 mM Mg2+, 50 μM ATP-Mg2+, and 2 μM CYT-19.
Using this substrate, we measured ATPase activity by CYT-19 under defined conditions (10 mM Mg2+; Fig. 1B). Nearly all of the ATPase activity arose from interactions with the duplex under these conditions, because the rate was ≈10-fold lower in the presence of either strand alone (Fig. 1B). Because the duplex separation reactions included a small excess of CCCUCUA5 and were performed under subsaturating conditions, we subtracted from the total the rate in the presence of 1 μM CCCUCUA5, which approximates its free concentration in reactions including the duplex. After subtracting this background, we obtained a duplex-dependent rate of 0.74 ± 0.05 μM/min (Fig. 1B and Table 1).
Table 1.
ATP utilization for CYT-19-mediated separation of the 6-bp P1 duplex
| [Mg2+], mM | Total ATPase rate | Duplex-dependent ATPase rate* | Total strand separation rate | CYT-19-dependent strand separation rate† | ATP hydrolyzed per strand separation |
|---|---|---|---|---|---|
| 2 | 1.10 ± 0.03 | 0.50 ± 0.03 | 1.05 ± 0.05 | 0.97 ± 0.05 | 0.45 ± 0.04 |
| 5 | 1.25 ± 0.04 | 1.10 ± 0.05 | 1.25 ± 0.10 | 1.20 ± 0.10 | 0.90 ± 0.09 |
| 10 | 0.80 ± 0.05 | 0.74 ± 0.05 | 0.73 ± 0.05 | 0.69 ± 0.05 | 1.1 ± 0.1 |
| 20 | 0.13 ± 0.01 | 0.11 ± 0.01 | 0.10 ± 0.01 | 0.05 ± 0.01 | 2.0 ± 0.4 |
Reactions were performed at 25°C with 50 mM Na-MOPS (pH 7.0), 50 μM ATP-Mg2+, 0.5 μM duplex, 1.2 μM total CCCUCUA5, 2 μM CYT-19. All rates are micromolar per minute. Values are averages and standard deviations from 2 to 4 independent determinations.
*Values are the total ATPase rate minus the ATPase rate in the presence of the approximate concentration of excess CCCUCUA5 expected to be single-stranded (0.7–1 μM). This difference represents the rate of ATPase activity arising from CYT-19 interacting with the duplex. The background ATPase activity was measured in the presence of 1 μM CCCUCUA5 to allow for the possibility of incomplete duplex formation, and control reactions established that the background rate depends only weakly on CCCUCUA5 concentration between 0.7 and 1 μM (<20% at 2 mM Mg2+ and <10% at 10 mM Mg2+; data not shown).
†Values are the observed rate constant for strand separation minus the basal strand separation rate constant in the absence of CYT-19, with the difference multiplied by the duplex concentration (0.5 μM) to give a steady-state rate.
We then measured P1 duplex separation under the same conditions, using a pulse–chase gel mobility shift assay (Fig. 1C and Fig. S2). After subtracting the CYT-19-independent separation, we obtained a rate constant of 1.4 ± 0.1 min−1 from a fit by a first-order rate equation (Fig. 1C and additional replicates not shown). To compare the rate of strand separation with the steady-state rate of ATPase activity measured above, we converted the rate constant to a steady-state rate by multiplying it by the duplex concentration (see SI Text, Analysis of Strand Separation). This conversion gave a steady-state rate of 0.69 ± 0.05 μM/min (Table 1). Strikingly, the rates of ATPase activity and strand separation are the same within error, giving a ratio of 1.1 ± 0.1 ATP molecules hydrolyzed per duplex separated. This ratio, or ATP utilization value, was the same within error across the range of experimentally accessible ATP concentrations (5–150 μM, data not shown; KM = 200 μM, ref. 2) and across the more limited range of accessible CYT-19 concentrations (0.5–2 μM, data not shown).* Thus, under these conditions (10 mM Mg2+, 25 °C), a single cycle of ATP-dependent conformational changes apparently gives complete strand separation of this 6-bp duplex. If any of the base pairs are not disrupted directly by the enzyme during this cycle, they must dissociate spontaneously.
Enhancement of Strand Separation by Bound ATP Without Hydrolysis.
To explore whether hydrolysis of ATP is uniformly required for strand separation by DEAD-box proteins, we decreased the Mg2+ concentration, shown previously to increase the strand separation activity of CYT-19 (4, 6). With 5 mM Mg2+, the ATP utilization value remained ≈1 (Table 1). With lower Mg2+ concentration (2 mM), however, it decreased to 0.45, implying that approximately half of the strand separation events proceeded without ATP hydrolysis. ATP-independent separation of this duplex has been shown for the related DEAD-box protein Mss116p (18), and we confirmed that, with 2 mM Mg2+ but not 10 mM Mg2+, CYT-19 gives detectable strand separation in the absence of nucleotide (with a rate of 0.2 μM/min; Fig. S3). This activity may be achieved by “strand capture,” analogous to the RNA chaperone activity of proteins that are not ATPases (18–20).
We therefore considered a model in which the ATP utilization value of 0.45 reflected a balance between strand separation mediated by ATP-bound CYT-19, which would result in ATP hydrolysis, and nucleotide-free CYT-19, which would of course not give ATP hydrolysis. However, it was not clear that the unwinding rate in the absence of ATP (0.2 μM/min; above) was large enough to support this model. In order for half of the separation events in the presence of ATP to be mediated by nucleotide-free CYT-19, the ATP-independent rate would have to be at least half of the total CYT-19-dependent rate of 0.97 ± 0.05 μM/min (Table 1). Although the difference between the expected and observed values is not large, it raised the possibility that bound ATP stimulates the rate of strand separation by CYT-19 even when it is not hydrolyzed.
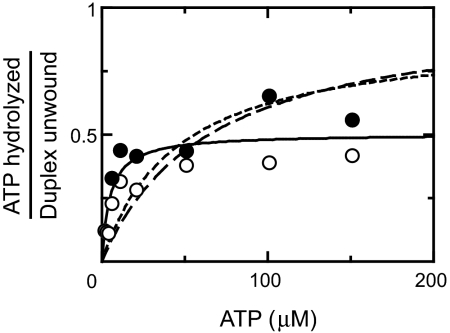
We tested this possibility by systematically varying the ATP concentration under the same 2 mM Mg2+ conditions (Fig. 2). If the low ATP utilization value resulted from activity of nucleotide-free CYT-19, it would increase with increasing ATP concentration, reflecting the increased fraction of CYT-19 bound to ATP, and would approach unity with saturating ATP (see SI Text, Stimulation of Strand Separation by Bound ATP and Scheme S1). Instead, the ATP utilization reached a plateau value of 0.5–0.6, giving no further increase with ATP concentration, even as the rate of strand separation increased to a value ≥40-fold larger than without ATP. This behavior indicates that ATP is hydrolyzed only in approximately half of the strand separation events, even when it is bound, and that bound ATP strongly accelerates strand separation even when it is not hydrolyzed (≥20-fold)†. These results strongly suggest that ATP elicits or stabilizes a protein conformation that induces or captures local strand separation events (see below and Fig. 3).
Fig. 2.
ATP hydrolyzed by CYT-19 per separation event of the 6-bp P1 duplex with low Mg2+ concentration (2 mM). Open and filled circles show results from 2 identical experiments. The solid curve shows the best fit by a model that includes stimulation of unwinding by bound ATP without hydrolysis (see SI Text, Stimulation of Strand Separation by Bound ATP). Dashed curves show best fits to discarded models in which only free CYT-19 gives ATP hydrolysis-independent strand separation (long dashes) or in which ATP-bound CYT-19 can give strand separation without hydrolysis, but with the same efficiency as nucleotide-free CYT-19 (short dashes).
Fig. 3.
Model for duplex separation by DEAD-box proteins. Interactions are formed between the ATP-bound helicase core and the RNA (radiolabeled RNA indicated by an asterisk), and a tethering interaction is formed adjacent to the core by an ancillary site, as shown, or by an additional protomer (7). Concomitant with or subsequent to initial binding by the helicase core, a conformation that depends on ATP binding but not hydrolysis induces or captures local strand separation. Complete strand separation can be achieved without ATP hydrolysis (Left, shaded blue) or with ATP hydrolysis (pathways down and to the right), which accelerates dissociation of the helicase core and may induce additional strand separation. Premature dissociation of the helicase core after ATP hydrolysis leads to a futile cycle (counterclockwise within box). Throughout the entire strand separation process, the tethering interaction may remain intact, as shown, allowing the protein to perform multiple cycles of structure disruption on the same RNA without being lost to solution.
Interestingly, we found that the enhancement from bound ATP is not mimicked by the nonhydrolyzable analog AMP-PNP, which gave no acceleration of strand separation beyond the basal level in the absence of nucleotide (Fig. S3). The lack of activity is not due to a failure to bind CYT-19, because AMP-PNP inhibited ATP stimulation with a KI of 200 μM (data not shown), 5-fold lower than the concentration used to test for stimulation. Thus, these results suggest that AMP-PNP binding does not elicit the same conformation as ATP, a conclusion that is strongly supported by recent work with a series of nucleotide analogs (21).
Increased ATP Requirement for Longer or More Stable Duplexes.
Regardless of the mechanism of strand separation, it would be expected that more ATP hydrolysis would be necessary for longer duplexes. An increased ATP requirement is further suggested for DEAD-box proteins by previous observations that longer duplexes are separated with greatly reduced rates. However, to our knowledge, the dependence of duplex length on the ATPase activity of DEAD-box proteins has not been systematically investigated.
Therefore, we extended the 5′-end of the hybrid strand with uridines to form P1 duplexes of 7, 9, or 11 base pairs (see Fig. 1A and Fig. S4). As expected, the ATP requirement increased with duplex length (Table 2). This increase arose from a decreased strand separation rate, whereas the ATPase rate remained essentially constant (Table S1). The insensitivity of the ATPase rate suggests a simple model in which CYT-19 manipulates both the longer and shorter duplexes by using a single cycle of ATP-dependent conformational changes to induce local strand separation, but for the longer duplexes a fraction of these events do not lead to complete strand separation, allowing rezipping of the duplex after the core domain of CYT-19 dissociates (see Fig. 3 and refs. 10, 22, and 23). These experiments also established 11 base pairs as a lower limit on the unwinding that can be accomplished using a single ATP, because this duplex gave an ATP utilization value of 1.2 ± 0.1 (2 mM Mg2+).
Table 2.
Dependence of ATP utilization by CYT-19 on Mg2+ concentration and duplex length
| [Mg2+], mM | Duplex length |
|||
|---|---|---|---|---|
| 6 bp | 7 bp | 9 bp | 11 bp | |
| 2 | 0.45 ± 0.04 | 0.4 ± 0.1 | 1.1 ± 0.1 | 1.2 ± 0.1 |
| 5 | 0.90 ± 0.09 | 1.3 ± 0.4 | 1.4 ± 0.4 | 4.4 ± 0.7 |
| 10 | 1.1 ± 0.1 | 1.3 ± 0.2 | 5 ± 2 | 11 ± 2 |
| 20 | 2.0 ± 0.4 | 2.0 ± 0.4 | 14 ± 2 | 18 ± 2 |
Values are ATP hydrolyzed per duplex separated (ATP utilization value). Longer duplexes were derived from that shown in Fig 1A by extending the 5́-end of the RNA/DNA strand with uridine nucleotides. All reactions were performed at 25°C with 50 mM Na-MOPS (pH 7.0), 50 μM ATP-Mg2+, 0.5 μM duplex, and 2 μM CYT-19.
We next increased the duplex stability, without increasing the length, by changing the natural G·U wobble pair within the P1 duplex to a G-C pair (Fig. S4). The G·U pair has been shown to destabilize the P1 helix, relative to the G-C, in part by disrupting base stacking (24, 25). We tested the effects of this base pair in the context of the 6-bp and 11-bp duplexes. Under standard conditions (10 mM Mg2+), the ATP utilization value for the 6-bp duplex increased 8-fold (from 1.1 ± 0.1 to 9 ± 3, Table S2), and in the context of the 11-bp duplex it increased 5-fold (from 11 ± 2 to 56 ± 11). Increases were also observed at lower Mg2+ concentrations, although the changes were smaller (2–4-fold) (Table S2). These results underscore the established links between duplex stability and the efficiency of separation by DEAD-box proteins (10), and they indicate that the less efficient unwinding for a more stable duplex does not simply result from slower action by DEAD-box proteins, but is accompanied by an increase in ATP consumption. This increase would not be expected for a conventional helicase and, as above, may reflect ATP hydrolysis events that are nonproductive because the core domain of CYT-19 dissociates before strand separation is complete (see Discussion).
A model involving nonproductive ATPase cycles would suggest that conditions that weaken CYT-19 binding or stabilize the duplex would give increased ATP requirements. Although changes in experimental conditions invariably give complex effects with multiple physical origins, changing Mg2+ concentration and temperature generally conformed to these expectations. With increased Mg2+ concentration, strand separation rates decreased substantially (Table 1 and Tables S1 and S2) and the ATP utilization values increased. These changes most likely include contributions from increased duplex stability (4, 26) and weaker binding by CYT-19 (S. Mohr and A.M.L, unpublished data). Lower temperatures also gave increased ATP requirements (Table S3), presumably in part from increased duplex stability.
Similar ATP Utilization by Other DEAD-Box Proteins.
To explore whether the insights obtained for CYT-19 extend to other DEAD-box proteins, we tested the Saccharomyces cerevisiae proteins Mss116p and Ded1p. Like CYT-19, these proteins separated a 9-base pair version of the P1 duplex at low Mg2+ concentrations with hydrolysis of at most a single ATP (Table 3). Further, both proteins gave an ATP utilization of <1 at 2 mM Mg2+, indicating that they, like CYT-19, are capable of strand separation without ATP hydrolysis. With increasing Mg2+ concentration, the ATP utilization by each protein increased, analogous to the behavior of CYT-19. Mss116p consistently hydrolyzed less ATP per separation event than did Ded1p or CYT-19, perhaps reflecting tighter binding of Mss116p to single-stranded RNAs and intermediates formed during strand separation (27).
Table 3.
ATP utilization by the DEAD-box proteins Mss116p and Ded1p
| [Mg2+], mM | Mss116p | Ded1p | CYT-19 |
|---|---|---|---|
| 2 | 0.5 ± 0.1 | 0.52 ± 0.09 | 1.1 ± 0.1 |
| 5 | 0.8 ± 0.3 | 1.5 ± 0.9 | 1.4 ± 0.4 |
| 10 | 0.8 ± 0.2 | 1.9 ± 0.5 | 5 ± 2 |
| 20 | 2.6 ± 0.5 | 6 ± 3 | 14 ± 2 |
Values are ATP hydrolyzed per duplex separated. Separation of a 9-bp duplex was monitored (construct 3 in Fig. S4). Conditions were identical to experiments for CYT-19 except that lower concentrations of proteins, substrates, and ATP were used to compensate for increased activity of these proteins. For experiments with Mss116p, the protein concentration was 100 nM, the duplex concentration was 25 nM, and the ATP concentration was 0.5–100 μM, depending on the Mg2+ concentration. For experiments with Ded1p, the protein concentration was 100–500 nM, the duplex concentration was 25–125 nM (maintaining a constant ratio of Ded1p and duplex concentrations), and the ATP concentration was 1–200 μM. In all cases, the ATP utilization values reported are saturating with respect to ATP concentration (data not shown). Corresponding values for CYT-19 are reproduced from Table 2 for comparison.
Discussion
By quantitatively comparing rates of ATPase activity and strand separation by the DEAD-box protein CYT-19 and related proteins, we have measured the number of ATP molecules hydrolyzed during separation of short helices under a range of solution conditions and temperatures (summarized in Table S4). Although analogous measurements have been made for several processive SF1 and SF2 helicases (reviewed in ref. 28), few have been reported for DEAD-box proteins (22), and none for DEAD-box proteins with short helices that are characteristic of structured RNAs. Our central conclusions are, first, that DEAD-box proteins can separate short duplexes in a single cycle of ATP-dependent conformational changes. Second, the process of strand separation is initiated, and sometimes even completed, while the proteins remain in the ATP-bound form. As described below, these insights provide critical new mechanistic information on strand separation by DEAD-box proteins, supporting and extending models for their action on physiological substrates (Fig. 3).
Strand Separation Depends on ATP Binding, Not Hydrolysis.
Upon binding an RNA duplex, a conformational change is suggested to result in tight binding of the protein to one strand of the RNA in a conformation that is incompatible with a duplex (Fig. 3 Upper Left). This conformational change depends on ATP binding but not hydrolysis, consistent with prior findings of cooperativity between binding of ATP and ssRNA (29, 30). It may induce local strand separation or trap a single-stranded segment that emerges due to transient “breathing” of the duplex. After this initial separation, which is suggested from structural analysis to be limited to 5 or 6 base pairs (31–33), additional base pairs can apparently dissociate spontaneously to allow complete separation of duplexes up to at least 9 base pairs in the absence of any ATP hydrolysis (pathway shaded blue in Fig. 3). The existence of this pathway is indicated by the ATP utilization values of <1.
The conclusion that nucleotide-dependent unwinding does not require hydrolysis is strongly supported by the finding from E. Jankowsky and colleagues that the nonhydrolyzable analog ADP-BeFx also promotes unwinding (21), and our demonstration that ATP gives this enhancement strongly supports the conclusion that ADP-BeFx provides a good approximate model for the ATP-bound state of DEAD-box proteins (21). Both studies also show that the enhancement by ATP is not mimicked by AMP-PNP. The Jankowsky group reports that unwinding of the longer duplexes in their study is undetectable with AMP-PNP, consistent with earlier work for several DEAD-box proteins and indicating that any stimulation by AMP-PNP is much less than that of ATP (12, 34, 35). Our work extends these results; because CYT-19-mediated separation of the 6-bp duplex can be monitored in the absence of any nucleotide, we are able to show that AMP-PNP provides no stimulation relative to this basal level. Whereas the lack of detectable activity with AMP-PNP has been suggested to indicate a requirement for ATP hydrolysis, the present results indicate that the defect arises at least in part from differences between the ATP-bound and AMP-PNP-bound states. A similar suggestion was made previously for the eIF4A protein from differences between ATP and AMP-PNP in RNA binding and cross-linking experiments (29). These results cast doubt on the general applicability of AMP-PNP as an analog of ATP for interactions with DEAD-box proteins. Although AMP-PNP can give tight binding of ssRNA to DEAD-box proteins (7, 21, 30, 31), this tight complex may not reflect an on-pathway intermediate for ATP-dependent strand separation. Alternatively, it may be on-pathway but attained with poor efficiency when starting from a double-stranded RNA and bound AMP-PNP.
What Is the Role of ATP Hydrolysis?
Although some strand separation occurs in the absence of ATP hydrolysis, the ATPase activities of all 3 DEAD-box proteins are substantially higher in the presence of a duplex than with either strand alone (Fig. 1 and data not shown). Thus, interactions with the duplex stimulate ATP hydrolysis, suggesting that ATP is sometimes hydrolyzed during strand separation. This ATP hydrolysis could occur from an intermediate complex in which both strands remain present, in which case it could give disruption of additional base pairs, or it could follow complete separation and function solely to facilitate dissociation of the helicase core from the tightly bound strand to allow additional cycles of unwinding (Fig. 3 and ref. 21).
Although the precise role remains an open question, it is tempting to suggest that ATP can be hydrolyzed before complete strand separation, as such premature hydrolysis could account for the observations by us and others that separation of longer duplexes is accompanied by hydrolysis of >1 ATP (22). If ATP were hydrolyzed before complete strand separation, premature dissociation of the helicase core could then allow rezipping of the duplex and “wasted” ATP hydrolysis (boxed pathway in Fig. 3). Supporting this interpretation, the ATPase rate remains constant as the length of the duplex increases, whereas the strand separation rate decreases (22), suggesting that the steps up to and including ATP hydrolysis are not affected by length, but that the longer duplexes are then less likely to become completely separated. It should be noted that alternative models for increased ATP utilization are possible, at least for longer duplexes where the participation of multiple functional units of protein could be imagined. However, even a duplex as short as 6-bp, which is almost certainly bound by only 1 monomer (31), can have an ATP requirement exceeding unity (Table S2). These results lead us to favor the nonproductive cycles shown in Fig. 3 as a central origin of the increased ATP requirements.
Notably, both the ATP-hydrolysis-dependent and -independent pathways can give complete strand separation in a single cycle, yielding ATP utilization values of 1 or lower and ruling out a general requirement for multiple cycles of ATP-hydrolysis-dependent translocation along the duplex (see SI Text, Distinct Mechanism from Processive Helicases). Importantly, the same general model could apply to tertiary contact disruption or protein displacement, because the critical feature is that a single strand of RNA is bound tightly and prevented from interacting with alternative partners.
Implications for Physiological Activities.
These mechanistic features of DEAD-box proteins are likely to be critical for their roles in manipulating structured RNAs and RNPs. A tethering interaction, which for CYT-19 and Mss116p appears to be mediated by the C-terminal domain (6, 36), positions the helicase core to disrupt nearby RNA structure. For CYT-19 and Mss116p, this interaction is relatively nonspecific, whereas for DEAD-box proteins that function with a defined RNA or RNP, this interaction is likely to be specific (12, 14). For both classes, it has been proposed that the tethering interaction may be maintained during local strand separation (4, 12). Here, we have tested this model and provide data in strong support of it. A single cycle of ATP-dependent conformational changes is sufficient to give complete disruption of short helices, and such a cycle can easily be envisioned to occur while the tethering interaction is maintained. This continued interaction may allow DEAD-box proteins to disrupt the same local structure repeatedly, which may be necessary to resolve misfolded RNAs such as group I introns (37), or to remain poised to facilitate rearrangements of newly formed intermediates that would otherwise revert rapidly to nonproductive structures. The same mechanism may also permit DEAD-box and related proteins that function in such processes as pre-mRNA splicing to mediate rapid and repeated interconversion of alternative sets of contacts, improving fidelity by allowing sampling of alternative conformations and intermolecular contacts (38–40).
Materials and Methods
Materials.
Oligonucleotides were purchased from Dharmacon. CCCUCUA5 was 5′-end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase and gel-purified (41). CYT-19, Mss116p, and Ded1p were expressed and purified as described (6, 27). AMP-PNP was treated to remove any contaminating ATP as described in ref. 18. RNA and nucleotide concentrations were determined spectrophotometrically (see SI Text, Determination of RNA and Nucleotide Concentrations).
RNA Strand Separation.
Unless otherwise indicated, reaction conditions were 25 °C, 50 mM Na-MOPS, pH 7.0, 10 mM MgCl2, 50 μM ATP-Mg2+, 50 mM KCl, and 5% glycerol. Reactions were initiated by adding preformed duplex (final concentrations of 0.5 μM RNA/DNA hybrid oligonucleotide, 0.2 μM CCCUCUA5, and trace 32P-labeled CCCUCUA5) to CYT-19 (2 μM), followed by addition of 1–5 μM unlabeled CCCUCUA5 to give a final duplex concentration of 0.5 μM. Control reactions showed that varying the concentration of unlabeled CCCUCUA5 across and beyond this range did not affect the rate constant for strand separation (Fig. S2), and thus 5 μM CCCUCUA5 was used typically to increase the signal for strand separation. At various times, aliquots were quenched by adding 70 mM MgCl2 and 1 mg/ml Proteinase K, and then loaded on a 20% nondenaturing polyacrylamide gel run at 5 °C. We confirmed that the quench solution was effective, because CYT-19 did not promote strand separation under the quench conditions (data not shown). Gels were dried, visualized with a phosphorimager (GE Healthcare), and quantitated with ImageQuant 5.2 (GE Healthcare). Time courses were fit by a single exponential equation (Kaleidagraph, Synergy Software). Rate constants were converted to steady-state rates by multiplying by the duplex concentration, 0.5 μM. A control experiment in which unlabeled chase CCCUCUA5 was added before labeled CCCUCUA5, such that CYT-19-mediated strand separation of the unlabeled P1 duplex was monitored by detecting the formation of labeled duplex, gave the same rate constant within error (data not shown). Thus, the presence of the 5′-phosphoryl group on the radiolabeled CCCUCUA5 does not affect the rate of CYT-19-mediated strand separation.
ATP Hydrolysis.
Conditions were as above except that reactions included trace [γ-32P]ATP instead of 32P-labeled CCCUCUA5. Unlabeled CCCUCUA5 was 1 μM (see above). Aliquots were quenched with 100 mM EDTA, applied to a polyethyleneimine (PEI) cellulose TLC plate, developed in 1 M formic acid, 0.5 M LiCl, and quantitated as above.
Supplementary Material
Acknowledgments.
We thank Eckhard Jankowsky, Dan Herschlag, and Sabine Mohr for comments on the manuscript. This work was supported by National Institutes of Health Grants GM070456 (to R.R.) and GM037951 (to A.M.L.) and Welch Foundation Grant F-1563 (to R.R.). M.D. is the recipient of National Institutes of Health Postdoctoral Fellowship F32-GM76961.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811075106/DCSupplemental.
The CYT-19 concentration can be varied only over a limited range with our current methods because at low concentrations the ATPase rate becomes too small to measure the fraction of ATP hydrolyzed, and at high CYT-19 concentrations, the strand separation becomes too fast for hand pipetting. Across the accessible range (0.5–2 μM, with CYT-19 in 4-fold excess of the duplex), the rate constant for strand separation increased approximately linearly, indicating that CYT-19 is subsaturating with respect to the duplex, and the ATP utilization value was unchanged within the expected limits of uncertainty (data not shown).
This value arises from the strand separation rate in the presence of ATP under conditions that do not favor its hydrolysis. The rate is 2–4 μM/min with 150 μM ATP, with no indication of saturation (data not shown), and therefore at least 8 μM/min with saturating ATP. ATP is hydorlyzed in only half of the strand-separation events. Thus, the pathway that involves bound ATP but not its hydrolysis must give half of the total rate (4 μM/min), ≥20-fold faster than CYT-19-dependent strand separation in the absence of ATP.
References
- 1.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 3.Huang HR, et al. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tijerina P, Bhaskaran H, Russell R. Non-specific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci USA. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci USA. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grohman JK, et al. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: The C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry. 2007;46:3013–3022. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 8.Jankowsky E, Fairman ME. RNA helicases—one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Rozen F, et al. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 11.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsu CA, Kossen K, Uhlenbeck OC. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExD/H protein YxiN is sufficient to confer specificity for 23S rRNA. J Mol Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 15.Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. YxiN is a modular protein combining a DEx(D/H) core and a specific RNA-binding domain. J Biol Chem. 2005;280:35499–35505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 16.Grifo JA, Abramson RD, Satler CA, Merrick WC. RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem. 1984;259:8648–8654. [PubMed] [Google Scholar]
- 17.Peck ML, Herschlag D. Effects of oligonucleotide length and atomic composition on stimulation of the ATPase activity of translation initiation factor eIF4A. RNA. 1999;5:1210–1221. doi: 10.1017/s1355838299990817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Del Campo M, et al. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 20.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA. doi: 10.1073/pnas.0811115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 23.Henn A, Cao W, Hackney D, De La Cruz EM. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J Mol Biol. 2008;365:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karbstein K, Lee J, Herschlag D. Probing the role of a secondary structure element at the 5′- and 3′-splice sites in group I intron self-splicing: The Tetrahymena L-16 ScaI ribozyme reveals a new role of the G. U pair in self-splicing. Biochemistry. 2007;46:4861–4875. doi: 10.1021/bi062169g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allain FH, Varani G. Structure of the P1 helix from group I self-splicing introns. J Mol Biol. 1995;250:333–353. doi: 10.1006/jmbi.1995.0381. [DOI] [PubMed] [Google Scholar]
- 26.Rogers GW, Jr, Lima WF, Merrick WC. Further characterization of the helicase activity of eIF4A. Substrate specificity. J Biol Chem. 2001;276:12598–12608. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 27.Halls C, et al. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 29.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 30.Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 31.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 32.Andersen CB, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 33.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Ray BK, et al. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- 35.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 36.Mohr G, et al. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J Mol Biol. 2008;375:1344–1364. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staley JP, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 39.Chen JY, et al. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol Cell. 2001;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 40.Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 41.Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: Enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.