Figure 2.
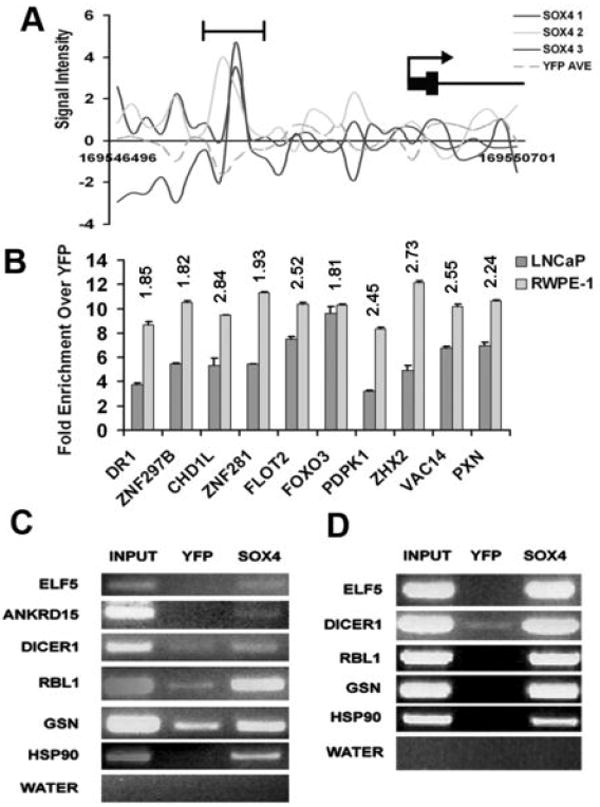
(A) Graph showing enrichment in the three HA-SOX4 lanes over the average of the two YFP replicates for the SOX4 target gene FMO4 Y-axis is the signal intensity across the genomic coordinates on the X-axis. (B) qPCR ChIP analysis of 10 randomly selected genes verified in both the RWPE-1 and LNCaP cell lines. Graph shows fold enrichment of the HA-SOX4 IP over the YFP negative control IP. Numbers above the bars represent the mean log2 of fold enrichment of ChIP-chip signal for the probes contained in the peak relative to YFP. Error bars indicate 1 SD (n = 3 technical replicates). (C) and (D) Genes that were verified by conventional ChIP assay. HA-SOX4 and YFP cells were subjected to conventional ChIP followed by PCR in both the LNCaP (C) and RWPE-1 (D) prostate cell lines. Six genes verified in the LNCaP cell lines and five in the RWPE-1 cell lines.
