Abstract
The compartmentalization of eukaryotic cells requires that newly synthesized proteins be targeted to the compartments in which they function. In chloroplasts, a few thousand proteins function in photosynthesis, expression of the chloroplast genome, and other processes. Most chloroplast proteins are synthesized in the cytoplasm, imported, and then targeted to a specific chloroplast compartment. The remainder are encoded by the chloroplast genome, synthesized within the organelle, and targeted by mechanisms that are only beginning to be elucidated. We used fluorescence confocal microscopy to explore the targeting mechanisms used by several chloroplast proteins in the green alga Chlamydomonas. These include the small subunit of ribulose bisphosphate carboxylase (rubisco) and the light-harvesting complex II (LHCII) subunits, which are imported from the cytoplasm, and 2 proteins synthesized in the chloroplast: the D1 subunit of photosystem II and the rubisco large subunit. We determined whether the targeting of each protein involves localized translation of the mRNA that encodes it. When this was the case, we explored whether the targeting sequence was in the nascent polypeptide or in the mRNA, based on whether the localization was translation-dependent or -independent, respectively. The results reveal 2 novel examples of targeting by localized translation, in LHCII subunit import and the targeting of the rubisco large subunit to the pyrenoid. They also demonstrate examples of each of the three known mechanisms—posttranslational, cotranslational (signal recognition particle-mediated), and mRNA-based—in the targeting of specific chloroplast proteins. Our findings can help guide the exploration of these pathways at the biochemical level.
Keywords: chloroplast, FISH, organelle, protein targeting, translation
Each organelle in a eukaryotic cell requires a distinct protein complement to carry out its specialized functions. Therefore, newly synthesized proteins are targeted to specific organelles and compartments within them. This process is known to involve 3 general mechanisms. In a posttranslational mechanism, the import machinery of the organelle selects the correct proteins by virtue of their having a transit peptide or nuclear localization signal. In a cotranslational mechanism, the signal sequence in the nascent polypeptide binds signal recognition particles (SRPs), which represses further translation and docks the mRNA–ribosome–nascent polypeptide complex at the endoplasmic reticulum (ER), whereupon translation resumes for the insertion of the elongating polypeptide into the ER lumen or membrane. Finally, in an mRNA-based mechanism, the untranslated mRNA is localized by an RNA-binding protein (RBP) associated with a molecular motor or the target membrane, and translation is initiated only on mRNA localization (1, 2). The two latter mechanisms appear to operate together in protein targeting to the mammalian ER (3).
The chloroplasts of plants and green algae import a few thousand proteins to function in photosynthesis and other processes (4). These proteins are believed to be synthesized at random cytoplasmic locations and to undergo posttranslational import, because isolated chloroplasts can import proteins synthesized in vitro, and EM studies have found only outer chloroplast envelope membranes without bound ribosomes (5, 6). But at least a few proteins are trafficked to chloroplasts through the secretory system, implying they are first targeted to the ER by the cotranslational SRP pathway (7–9). Despite several decades of research on chloroplast protein import, the location of chloroplast protein synthesis in the cytoplasm remains unknown.
A minor protein contingent is expressed from the chloroplast genome and targeted to specific compartments within this organelle (10). Some of these “chloroplast-encoded” proteins are targeted to thylakoid membranes, where they function as subunits in the photosynthesis complexes. Others are targeted to the stroma. In algae, the rubisco large subunit (LSU) is targeted to the pyrenoid, a spherical chloroplast compartment specializing in CO2 fixation (11); however, little is known about the targeting mechanisms involved. We do know that thylakoid membrane proteins are synthesized by membrane-bound chloroplast polysomes (12, 13). A chloroplast homolog of the SRP subunit that binds the signal sequence, cpSRP54, has been shown to bind the nascent chain of the D1 subunit of photosystem II (PSII) in a reconstituted chloroplast translation system and to be required for thylakoid membrane biogenesis (14–17). mRNA-based mechanisms are thought to target chloroplast-encoded proteins as well, for 3 reasons. First, only one protein is known to have a transit peptide, indicating that posttranslational mechanisms are not predominant (14). Second, cpSRP54 is not essential for thylakoid biogenesis in Arabidopsis, indicating the existence of another pathway (18). Finally, several membrane-associated RBPs have been identified in the chloroplast of the eukaryotic green alga Chlamydomonas reinhardtii (19, 20). Much work remains to be done to identify and characterize the mechanisms and machinery involved in the targeting of chloroplast-encoded proteins.
To address these long-standing questions, we used in situ techniques in Chlamydomonas to identify the targeting mechanisms used by the following canonical chloroplast proteins: the rubisco small subunit (SSU), the light harvesting complex II (LHCII) subunits, and the chloroplast-encoded proteins D1 and LSU. Both mRNA-based and cotranslational targeting involve the localization of the mRNA encoding the targeted protein. Therefore, to distinguish these mechanisms from posttranslational targeting, we used FISH, immunofluorescence staining, and confocal microscopy to determine whether the mRNA is localized near the target compartment of the protein that it encodes. Cotranslational targeting requires translation to produce a signal peptide, whereas mRNA-based targeting does not, because the localization signal is in the mRNA. Therefore, we distinguished cotranslational and mRNA-based targeting based on whether or not pharmacological inhibition of translation prevents localization of the mRNA. Our results reveal the use of each of these 3 mechanisms in chloroplast protein targeting, along with 2 novel examples involving localized translation.
Results
LSU Targeting to the Pyrenoid Involves a Cotranslational Mechanism.
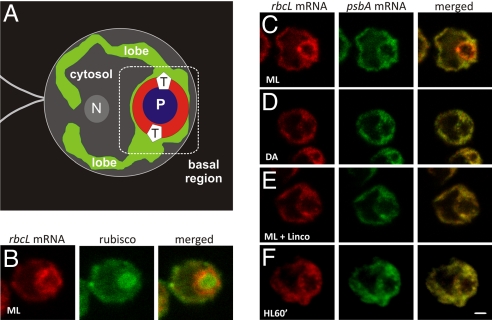
To characterize the targeting of LSU to the pyrenoid, we used FISH to analyze the distribution of its mRNA, that of the chloroplast rbcL gene. The rbcL mRNA signal was enriched in the chloroplast basal region relative to the lobes (Fig. 1A–C). Coimmunostaining of these cells for rubisco revealed that the rbcL mRNA was distinctly focused at the outer perimeter of the pyrenoid in most cells (see Materials and Methods). This localization pattern was not observed in the FISH signals of 3 chloroplast mRNAs encoding thylakoid membrane proteins: psbA (Fig. 1C), psbC, and psaA (21). Therefore, these results support the targeting of LSU to the pyrenoid by either a cotranslational or an mRNA-based mechanism.
Fig. 1.
Translation-dependent localization of the chloroplast rbcL mRNA at the pyrenoid for LSU targeting. (A) An illustration of the cell shown in (C) demonstrating the single chloroplast (green) with its lobes and globular basal region. The basal region contains the pyrenoid (P; blue). Also indicated are the cytosolic region (gray) and the approximate locations of T zones (T), the nucleus (N), and the flagella. The region in which rbcL mRNAs localize is shown in red. (B) An ML cell showing the rbcL mRNA signal and immunolabeled rubisco. (C–F) Signals from the rbcL and psbA mRNAs in cells from constant ML (C), DA (D), ML with lincomycin (E), and HL (F) for 60 min. The punctate colocalized psbA and rbcL mRNA signals near the pyrenoid in (E) and (F) are chloroplast stress granules, which form in response to oxidative stress caused by a secondary effect of lincomycin (photosensitization to ML) or HL, respectively (21). The micrographs show 0.2-μm optical sections. For each experiment, n ≥ 20 cells. Each pattern was observed in ≥ 80% of cells. (Scale bars: 1 μm.)
To determine whether these rbcL mRNAs are localized by targeting signals in the LSU nascent chain (in a cotranslational mechanism) or in the mRNA sequence (in an mRNA-based mechanism), we explored whether this localization occurs in cells depleted of translating chloroplast ribosomes and LSU nascent chains through treatment with lincomycin (22, 23). The rbcL FISH signal was not localized around the pyrenoid in cells treated with this drug (Fig. 1E). This localization pattern also was not seen in cells under conditions that down-regulate rbcL translation, a 2-h dark adaption (DA) (24, 25) or a 60-min high-light (HL) exposure (26) (Fig. 1 D and F). The apparent higher level of the rbcL FISH signal in cells exposed to moderate light (ML) compared with DA cells and lincomycin-treated ML cells most likely reflects the concentration of the signal near the pyrenoid; 4 previous studies found a constant rbcL mRNA level across diverse light conditions in Chlamydomonas (24, 25, 27, 28). Together, these results strongly support LSU targeting to the pyrenoid primarily through a cotranslational mechanism.
In a minority of cells (25%; n = 24), rubisco and the rbcL mRNA also colocalized in a structure with the size, shape, and general location of the eyespot (Fig. 1B). Because this localization pattern was not detected in most cells, we did not explore it further.
D1 Targeting for De Novo PSII Assembly Involves an mRNA-Based Mechanism.
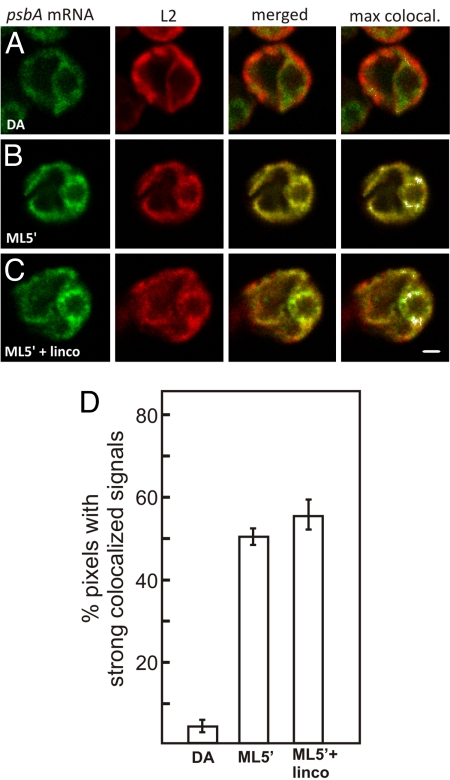
We also explored how D1 is targeted for de novo PSII assembly in T zones. T zones are punctate regions adjacent to the pyrenoid in which we previously observed the colocalization of multiple components in PSII subunit synthesis for de novo assembly when it was induced by shifting cells from darkness to ML for 5 min (ML5′ cells) (Fig. 1A) (29). Fig. 2A and B show the colocalization of the psbA mRNA and the chloroplast ribosomal protein L2 in T zones only in the inducing ML5′ condition, not in the repressing DA condition (see Materials and Methods). To highlight this colocalization pattern, the pixels with the strongest colocalized signals are displayed in white using ImageJ (30), as shown in the fourth image column in Fig. 2A–C (see also Fig. 3A–E). T zones are distinct from the rbcL mRNA localization pattern around the pyrenoid; they are punctate and located on the lateral sides of the pyrenoid, and the rbcL mRNA does not localize to them (29). Because the scoring of this pattern is subjective, we analyzed each T zone for the percentage of pixels with strong colocalized signals and determined the mean value for each condition (Fig. 2D) (29). These results confirmed our previous finding that the psbA mRNA localizes to T zones during the induction of de novo PSII assembly. Therefore, this targeting of D1 involves either a cotranslational or an mRNA-based mechanism.
Fig. 2.
Translation-independent localization of the psbA mRNA in T zones for de novo PSII assembly. (A–C) Fluorescence signals from the psbA mRNA and the chloroplast ribosomal protein L2 in cells from the following conditions: 2-h DA (A), after a 5-min ML exposure to initiate psbA translation for de novo PSII assembly (ML5′) (B), and ML5′ cells generated in the presence of lincomycin (C). The fourth image column shows the merged channels, with the strongest colocalized signals highlighted in white. The punctate psbA mRNA signal near the pyrenoid that does not colocalize with L2 is in chloroplast stress granules (21). The micrographs show 0.2-μm optical sections. (Scale bar: 1 μm.) (D) The percentage of pixels in sampled T zones with strong colocalized signals [white pixels in (A–C)] for each of the 3 conditions. The error bars indicate 2 standard errors. For each experiment, n ≥ 20 cells.
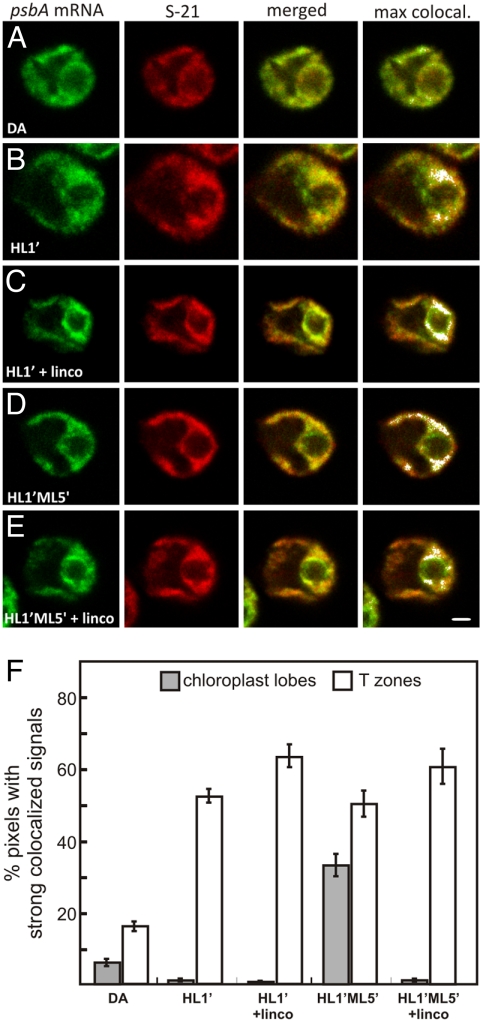
Fig. 3.
Localization of the psbA mRNA for the de novo assembly and repair of PSII. (A–E) Fluorescence signals from the psbA mRNA and the chloroplast ribosomal protein S-21 in cells from the following conditions: a 2-h DA (A); DA cells exposed to HL for 1 min (HL1′) to induce psbA translation for de novo PSII assembly (B); HL1′ cells generated in the presence of lincomycin (C); HL1′ cells exposed to ML for 5 min, a condition of PSII repair (HL1′ML5′) (D); and HL1′ML5′ cells generated in the presence of lincomycin (E). The micrographs show 0.2-μm optical sections. (Scale bar: 1 μm.) (F) The percentages of pixels with strong colocalized signals in T zones (white bars) and chloroplast lobes (shaded bars) across all cells from the 5 conditions. The error bars indicate 2 standard errors. For each experiment, n ≥ 20 cells.
To discriminate between these 2 mechanisms, we investigated whether or not the psbA mRNA localized to T zones after chloroplast mRNAs were cleared of ribosomes and nascent polypeptides by treatment with lincomycin. When ML5′ cells were generated in the presence of lincomycin, the psbA mRNA and L2 still colocalized in T zones (Fig. 2B and C). Moreover, the quantitative analyses revealed that lincomycin unexpectedly enhanced this colocalization (Fig. 2D). Lincomycin also enhanced colocalization of the psbA mRNA and a marker protein for the small chloroplast ribosomal subunit, S-21, under similar conditions that activated psbA translation for the de novo PSII assembly: DA cells exposed to HL for 1 min (HL1′ cells) (Fig. 3A–C and F). This translation-independence supports a primary mRNA-based mechanism in psbA mRNA localization to T zones for D1 synthesis in the de novo PSII assembly.
Comparing the distributions of L2 and S-21 revealed that chloroplast ribosomal subunits also localized to T zones through translation-independent mechanisms (Figs. 2 and 3).
D1 Targeting to Thylakoids for PSII Repair Involves a Cotranslational Mechanism.
During HL stress, the replacement of photodamaged D1 subunits in PSII complexes involves the localization of the psbA mRNA to thylakoid membranes throughout the chloroplast (29, 31). As a condition to induce PSII repair, we used HL1′ cells incubated for an additional 5 min under ML (HL1′ML5′), for reasons described in our previous report (29). This effect can be seen as the appearance of strong colocalized signals from the psbA mRNA and S-21 in chloroplast lobes in HL1′ML5′ cells (Fig. 3D) relative to HL1′ cells (Fig. 3B; in 3F, compare the second and fourth shaded bars). This localization of the psbA mRNA for D1 targeting in PSII repair is consistent with both cotranslational and mRNA-based mechanisms. To discriminate between these possibilities, we generated HL1′ML5′ cells in the presence of lincomycin. Under these conditions, we did not detect this colocalization of the psbA mRNA and L2 in the lobes, thus supporting a cotranslational mechanism in D1 targeting for PSII repair in thylakoid membranes (Fig. 3C and E; in 3F, compare the third and fifth shaded bars).
Chloroplast Protein Targeting From the Cytoplasm.
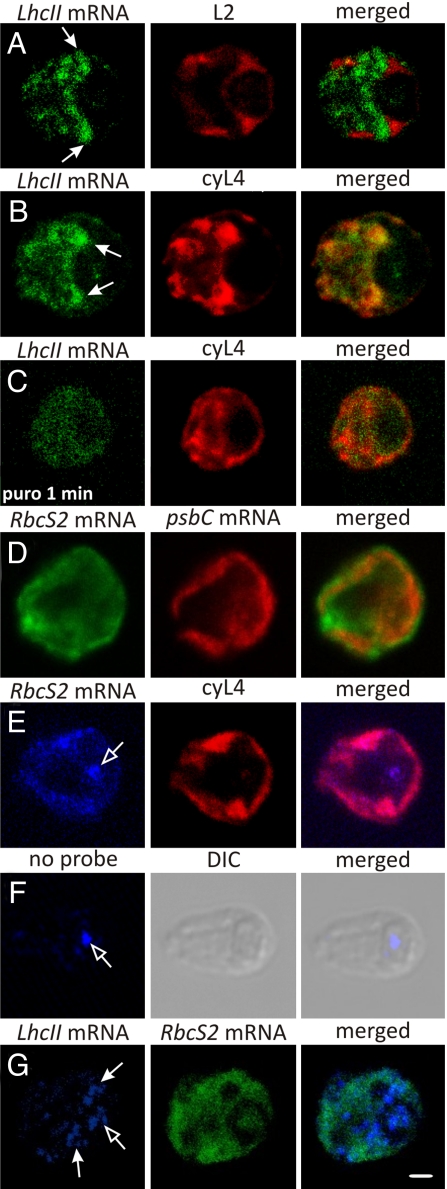
We then turned our attention to protein targeting from the cytoplasm to the chloroplast. Using FISH to characterize the distribution of LhcII mRNAs, which encode the subunits of LHCII (32), revealed that their FISH signal was frequently concentrated along the cytoplasmic border of the chloroplast basal region (see Materials and Methods). This localization pattern is shown in Fig. 4A, in which the chloroplast was stained with immunolabeled L2. In other cells, the LhcII mRNAs that localized at the basal region were more concentrated near the bases of chloroplast lobes (Fig. 4B and G). These results raise the possibility that the cytoplasmic border of the chloroplast basal region is a privileged location of LHCII synthesis. This hypothesis is supported by several findings. First, different distributions were observed for the RbcS2 mRNA (Fig. 4D–G; see the next paragraph) and the mRNA encoding β2-tubulin (33). Second, the localized LhcII mRNAs probably were translated, because they colocalized with the concentrated marker protein for cytoplasmic ribosomes, cyL4, and this LhcII mRNA localization pattern was not detected after mRNAs were released from ribosomes and nascent chains by puromycin (Fig. 4B and C, respectively) (12, 34). The concurrent decrease in the intensity of this signal likely reflects rapid degradation of untranslated LhcII mRNAs, because their translation and abundance are directly correlated in Chlamydomonas (27, 35). Third, patches of strong cyL4 signals at other cytoplasmic locations were not enriched in the LhcII mRNA signal (Fig. 4B); therefore, LHCII subunits appear to be targeted to the chloroplast by the localized translation of the mRNAs encoding them. The translation-dependence of this mRNA localization pattern supports a cotranslational mechanism.
Fig. 4.
FISH analyses of 2 nucleocytosolic mRNAs. (A) An ML cell showing the distribution of the LhcII mRNAs relative to the chloroplast, stained by immunolabeling L2. The closed-head arrows indicate colocalization near the chloroplast basal region. (B) An ML cell showing the fluorescence signals from LhcII mRNAs and the cytoplasmic ribosomal protein cyL4. (C) An ML cell exposed to puromycin for 1 min that was FISH-probed for the LhcII mRNAs and immunostained for cyL4. (D) An ML cell showing the distribution of the RbcS2 mRNAs relative to the chloroplast, which was FISH-probed for the psbC mRNA. (E) An ML cell that was FISH-probed for the RbcS2 mRNA and immunostained for the cytoplasmic ribosomal protein cyL4. The open-head arrow shows the autofluorescence resulting from excitation at 633 nm, seen in the nonprobed/immunostained cell in (F). (G) An ML cell that was FISH-probed for LhcII and RbcS2 mRNAs. DIC, differential interference contrast. The micrographs show 0.2-μm optical sections. (Scale bar: 1 μm.)
Analysis of the distribution of the RbcS2 mRNA, which encodes SSU, revealed that its FISH signal was not enriched at the chloroplast perimeter (Fig. 4D, E, and G). The prominent nonstaining region contains the nucleus (33). Some cells exhibited variable intensity in this signal, but this was not concentrated in any consistent location (Fig. 4D). This broad distribution of the RbcS2 mRNA was quite distinct from the LhcII mRNA localization pattern when these signals were compared in the same cells (Fig. 4G). Moreover, the RbcS2 mRNA colocalized with cyL4 throughout the cytosolic region (Fig. 4E). Therefore, these results support the current model of SSU synthesis throughout the cytoplasm and posttranslational import into the chloroplast.
Discussion
Our results provide evidence of chloroplast protein targeting by the 3 general mechanisms described earlier and reveal a remarkably complex spatial organization of chloroplast protein synthesis. The colocalization of RbcS2 mRNA with ribosomes throughout the cytosol provides the first in situ evidence supporting the long-standing model of SSU synthesis at random cytoplasmic locations and posttranslation import into the chloroplast (Fig. 4E) (4, 36). Although our findings do not exclude the possibility of a minor RbcS2 mRNA pool localized at the chloroplast perimeter for translation and SSU targeting, this seems unlikely, because confocal microscopy is sufficiently sensitive and quantitative for detecting such a pool. Most RbcS2 mRNAs likely are translated; their FISH signal was completely colocalized with the cyL4 signal (Fig. 4E). Moreover, SSU is among the most highly synthesized proteins in Chlamydomonas, and nearly all RbcS mRNAs are polysome-associated in barley leaf cells (36, 37).
In contrast with the nonlocalization of the RbcS2 mRNA, we found evidence for localized LhcII translation at the cytoplasmic border of the chloroplast basal region (Fig. 4A, B, and G). This could localize the import of newly synthesized LHCII subunits near T zones to facilitate the assembly of the PSII–LHCII supercomplex. To what feature of cellular architecture could LhcII mRNAs be localized? LHCII subunits likely are cotranslationally inserted into a membrane, because they are hydrophobic integral membrane proteins. They probably are not routed to the chloroplast through the secretory system in Chlamydomonas as they are in other algal species, because these proteins are predicted to lack a signal peptide by TargetP (38). Our working hypothesis is that polysomes with LhcII mRNAs are bound to specific regions of chloroplast envelope for cotranslational targeting of LHCII subunits to the chloroplast by unknown mechanisms. Although this hypothesis is seemingly contradicted by many EM images of ribosome-free outer chloroplast envelope membranes (5, 6, 39), none of these images were obtained from cells treated with an inhibitor of translation elongation, which is required to retain chloroplast ribosomes on thylakoid membranes during cell isolation (40).
We expanded our survey of mRNA localization patterns in the Chlamydomonas chloroplast by demonstrating that the rbcL mRNA is translated at the outer perimeter of the pyrenoid, probably to target LSU for the rubisco assembly therein (Fig. 1B and C) (11). Weaker rbcL mRNA signals were detected throughout the chloroplast, possibly from a pool translated for the rubisco bound to thylakoid membranes (41). The translation-dependence of the former localization pattern supports a cotranslational targeting mechanism. We also found similar evidence for cotranslational targeting of D1 for the repair of photodamaged PSII complexes in thylakoid membranes throughout the chloroplast (Fig. 3D–F). Future research should explore whether LSU and D1 targeting involves the cpSRP54 homolog in Chlamydomonas (AF238499).
An mRNA-based mechanism in D1 targeting for de novo PSII assembly is supported by the translation-independence of psbA mRNA localization to T zones (Figs. 2 and 3). The possibility exits that cpSRP54 subsequently directs nascent D1 for cotranslational membrane insertion. Concerted mRNA-based and cotranslational mechanisms have been speculated to operate in protein targeting to thylakoids, and there is evidence for this in protein targeting to the mammalian ER (14, 42).
We also found that chloroplast ribosomal subunits localized to T zones through translation-independent mechanisms. This finding was not unexpected, because 10%−40% of chloroplast ribosomes are associated with membranes by electrostatic interactions alone in Chlamydomonas and Pea (40, 43, 44). Moreover, translation-independent mechanisms localize ribosomes to the inner membrane of mitochondria in Saccharomyces cerevisiae and the mammalian ER (42, 45).
Our use of in situ approaches in the analysis of mRNA localization for chloroplast protein targeting has begun to identify the targeting mechanisms used by specific proteins and for different processes in the biogenesis and repair of the photosynthesis apparatus. We have found intriguing similarities to intracellular protein targeting in many other organisms. Although the posttranslational import machineries in the chloroplast envelope have been identified and are currently being dissected, much remains to be learned about the cotranslational and mRNA-based pathways (4). By revealing specific examples of chloroplast protein targeting by cotranslational and mRNA-based pathways, our results will help guide the exploration of these pathways at the biochemical level.
Materials and Methods
Culture Conditions.
Strain CC-503 was cultured in high-salt minimal medium (46) until the mid-log phase (ca. 3 × 106 cells mL−1) at 24 °C. ML was 100–150 μE m−2 s−1, and HL was 2,000 μE m−2 s−1. The deletion mutant for RbcS1 (and RbcS2), T60–3, was cultured on Tris-acetate-phosphate medium under indirect light (36). The conditions used (DA, ML, ML5′, HL1′, HL′ML5′) and inhibitor concentrations were described previously (21, 29).
FISH and Immunofluorescence Staining.
The FISH and immunofluorescence procedures, as well as the psbA and rbcL FISH probes, have been described previously (29, 33). The LhcII and RbcS2 FISH probes (Table 1) were labeled with either Alexa 488 or Alexa 633 (Molecular Probes) according to the manufacturer's protocol. The LhcII probes were designed to hybridize to the LhcII-3 mRNA (Accession BAB64417), but they also were complementary to the other 3 LhcII mRNAs. The RbcS2 probes hybridized to the RbcS2 mRNA (Accession P08475) and failed to produce a signal in a deletion mutant for both RbcS genes (Fig. S1A). The absence of the LhcII FISH signal in DA cells (Fig. S1B) provides negative controls for its specificity, in the absence of a deletion mutant for these genes (27). The antisera against the ribosomal proteins were described previously (29, 34, 46–49).
Table 1.
Oligonucleotide FISH probe sequences
| Probe sequence (5′ → 3′) | |
|---|---|
| RbcS-1 | GGGTTGCAAGTGCTCAAATACCCCATCAAACATCATCCTGGTTTGGCTGC |
| RbcS-2 | CGGAGGACTTGGCAATGACGGCGGCCATTTTAAGATGTTGAGTGACTTCT |
| RbcS-3 | TAGGAGAAGGTCTCGAACATCTTGACCGGGGTCCAGACCATCATCTGGTT |
| RbcS-4 | TTCCACATGGTCCAGTAGCGGTTGTCGTAGTACAGCTGCAAGACACGCTG |
| LhcII-1 | GCTGCGGACGGAGGACTTCATGATGGCGGCCATTTTGATTGGTATAGACA |
| LhcII-2 | AAGAAGCCGAACATGGAGAACATAGCCAGGCGGCCGTTCTTGATCTCCTT |
| LhcII-3 | TCTCCGCTCAATCACGCCAGTACATGCAGCTGCCGAGGGCCAAAAATTTA |
| LhcII-4 | TTAATCGCACGTCCCTCGCGCTCCTAAGCCTGTGAAAAGAGGCTCACACT |
Bold T's indicate modified C6-dT residues that were labeled with either Alexa Fluor 488 or Alexa Fluor 633 (Molecular Probes).
The localization patterns described herein were seen in 80% of the cells examined (n > 20), except in the following cases. In 55% of the cells, the rbcL mRNA was distinctly focused at the outer perimeter of the pyrenoid (Fig. 1C), whereas in 40% the signal was slightly more dispersed (Fig. 1B), and in 5% it was not localized (n = 20). An atypically high percentage of cells lacked the LhcII mRNA signal, for unknown reasons. Of the 110 ML cells examined, 62 (56%) had a very weak signal, which likely was background because it was evenly distributed throughout the cell, including within the chloroplast (data not shown). Considering just the 48 cells that had above-background LhcII FISH signals in the cytosolic region, 30 cells (63%) had the localization pattern shown in Fig. 4 A, B, and G, 14 cells (29%) had a less evident semblance of this localization pattern, and 4 cells (8%) had a nonlocalized signal.
Microscopy.
Fluorescence signals were visualized in 0.2-μm optical sections obtained with a Leica TCS SP2 confocal laser-scanning microscope and image acquisition software, version 2.61. Argon, green helium neon, and helium neon lasers were used to produce the 488-nm, 543-nm, and 633-nm stimulation of fluorophores Alexa 488/FITC, Alexa 555/TRITC, and Alexa 633, respectively. Cells were observed under settings described previously (21). Statistical analyses of colocalization were carried out as described previously (29).
Supplementary Material
Acknowledgments.
This study used the confocal microscope at Concordia University's Centre for Structural and Functional Genomics and was funded by Natural Sciences and Engineering Council of Canada Grant 217566–03. We thank D. Durnford, E. Schleiff, M. Champagne, M. Herrington, U. Oberholzer, and A. Piekny for stimulating discussions and manuscript review; E. Harris for antisera; and R. Spreitzer for the RbcS deletion mutant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811268106/DCSupplemental.
References
- 1.St. Johnston D. Moving messages: The intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 2.Sanchirico ME, Fox TD, Mason TL. Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J. 1998;17:5796–5804. doi: 10.1093/emboj/17.19.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephens SB, et al. Analysis of mRNA partitioning between the cytosol and endoplasmic reticulum compartments of mammalian cells. Methods Mol Biol. 2008;419:197–214. doi: 10.1007/978-1-59745-033-1_14. [DOI] [PubMed] [Google Scholar]
- 4.Inaba T, Schnell DJ. Protein trafficking to plastids: One theme, many variations. Biochem J. 2008;413:15–28. doi: 10.1042/BJ20080490. [DOI] [PubMed] [Google Scholar]
- 5.Chua NH, Schmidt GW. Transport of proteins into mitochondria and chloroplasts. J Cell Biol. 1979;81:461–483. doi: 10.1083/jcb.81.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carde JP, Joyard J, Douce R. Electron microscopic studies of envelope membranes from spinach plastids. Biol Cell. 1982;44:315–324. [Google Scholar]
- 7.Nanjo Y, et al. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell. 2006;18:2582–2592. doi: 10.1105/tpc.105.039891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Villarejo A, et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat Cell Biol. 2005;7:1224–1231. doi: 10.1038/ncb1330. [DOI] [PubMed] [Google Scholar]
- 9.Radhamony RN, Theg SM. Evidence for an ER to Golgi to chloroplast protein transport pathway. Trends Cell Biol. 2006;16:385–387. doi: 10.1016/j.tcb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Zerges W. Translation in chloroplasts. Biochimie. 2000;82:583–601. doi: 10.1016/s0300-9084(00)00603-9. [DOI] [PubMed] [Google Scholar]
- 11.Borkhsenious ON, Mason CB, Moroney JV. The intracellular localization of ribulose-1,5-bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. Plant Physiol. 1998;116:1585–1591. doi: 10.1104/pp.116.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chua NH, Blobel G, Siekevitz P, Palade GE. Periodic variations in the ratio of free to thylakoid-bound chloroplast ribosomes during the cell cycle of Chlamydomonas reinhardtii. J Cell Biol. 1976;71:497–514. doi: 10.1083/jcb.71.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Margulies MM. Synthesis of photosynthetic membrane proteins directed by RNA from rough thylakoids of Chlamydomonas reinhardtii. Eur J Biochem. 1983;137:241–248. doi: 10.1111/j.1432-1033.1983.tb07821.x. [DOI] [PubMed] [Google Scholar]
- 14.Eichacker LA, Henry R. Function of a chloroplast SRP in thylakoid protein export. Biochim Biophys Acta. 2001;1541:120–134. doi: 10.1016/s0167-4889(01)00151-3. [DOI] [PubMed] [Google Scholar]
- 15.Gutensohn M, et al. Toc, Tic, Tat, et al.: Structure and function of protein transport machineries in chloroplasts. J Plant Physiol. 2006;163:333–347. doi: 10.1016/j.jplph.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Aro EM. Synthesis, membrane insertion and assembly of the chloroplast-encoded D1 protein into photosystem II. FEBS Lett. 2002;512:13–18. doi: 10.1016/s0014-5793(02)02218-4. [DOI] [PubMed] [Google Scholar]
- 17.van Wijk KJ, Knott TG, Robinson C. Evidence for SecA- and delta pH-independent insertion of D1 into thylakoids. FEBS Lett. 1995;368:263–266. doi: 10.1016/0014-5793(95)00668-y. [DOI] [PubMed] [Google Scholar]
- 18.Amin P, et al. Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol. 1999;121:61–70. doi: 10.1104/pp.121.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nickelsen J. Chloroplast RNA-binding proteins. Curr Genet. 2003;43:392–399. doi: 10.1007/s00294-003-0425-0. [DOI] [PubMed] [Google Scholar]
- 20.Zerges W, Wang S, Rochaix JD. Light activates binding of membrane proteins to chloroplast RNAs in Chlamydomonas reinhardtii. Plant Mol Biol. 2002;50:573–585. doi: 10.1023/a:1020246007858. [DOI] [PubMed] [Google Scholar]
- 21.Uniacke J, Zerges W. Stress induces the assembly of RNA granules in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol. 2008;182:641–646. doi: 10.1083/jcb.200805125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Paakkarinen V, van Wijk KJ, Aro EM. Biogenesis of the chloroplast-encoded D1 protein: Regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell. 2000;12:1769–1782. doi: 10.1105/tpc.12.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edhofer I, Muhlbauer SK, Eichacker LA. Light regulates the rate of translation elongation of chloroplast reaction center protein D1. Eur J Biochem. 1998;257:78–84. doi: 10.1046/j.1432-1327.1998.2570078.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee J, Herrin DL. Assessing the relative importance of light and the circadian clock in controlling chloroplast translation in Chlamydomonas reinhardtii. Photosynth Res. 2002;72:295–306. doi: 10.1023/A:1019881306640. [DOI] [PubMed] [Google Scholar]
- 25.Herrin DL, Michaels AS, Paul AL. Regulation of genes encoding the large subunit of ribulose-1,5-bisphosphate carboxylase and the photosystem II polypeptides D-1 and D-2 during the cell cycle of Chlamydomonas reinhardtii. J Cell Biol. 1986;103:1837–1845. doi: 10.1083/jcb.103.5.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shapira M, et al. Differential regulation of chloroplast gene expression in Chlamydomonas reinhardtii during photoacclimation: Light stress transiently suppresses synthesis of the rubisco LSU protein while enhancing synthesis of the PSII D1 protein. Plant Mol Biol. 1997;33:1001–1011. doi: 10.1023/a:1005814800641. [DOI] [PubMed] [Google Scholar]
- 27.Malnoe P, Mayfield SP, Rochaix JD. Comparative analysis of the biogenesis of photosystem II in the wild-type and Y-1 mutant of Chlamydomonas reinhardtii. J Cell Biol. 1988;106:609–616. doi: 10.1083/jcb.106.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breidenbach E, Jenni E, Boschetti A. Synthesis of two proteins in chloroplasts and mRNA distribution between thylakoids and stroma during the cell cycle of Chlamydomonas reinhardii. Eur J Biochem. 1988;177:225–232. doi: 10.1111/j.1432-1033.1988.tb14366.x. [DOI] [PubMed] [Google Scholar]
- 29.Uniacke J, Zerges W. Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell. 2007;19:3640–3654. doi: 10.1105/tpc.107.054882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with Image. J. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 31.Adir N, Shochat S, Ohad I. Light-dependent D1 protein synthesis and translocation is regulated by reaction center II: Reaction center II serves as an acceptor for the D1 precursor. J Biol Chem. 1990;265:12563–12568. [PubMed] [Google Scholar]
- 32.Minagawa J, Takahashi Y. Structure, function and assembly of photosystem II and its light-harvesting proteins. Photosynth Res. 2004;82:241–263. doi: 10.1007/s11120-004-2079-2. [DOI] [PubMed] [Google Scholar]
- 33.Colon-Ramos DA, et al. Asymmetric distribution of nuclear pore complexes and the cytoplasmic localization of beta2-tubulin mRNA in Chlamydomonas reinhardtii. Dev Cell. 2003;4:941–952. doi: 10.1016/s1534-5807(03)00163-1. [DOI] [PubMed] [Google Scholar]
- 34.Fleming GH, Boynton JE, Gillham NW. Cytoplasmic ribosomal proteins from Chlamydomonas reinhardtii: Characterization and immunological comparisons. Mol Gen Genet. 1987;206:226–237. doi: 10.1007/BF00333578. [DOI] [PubMed] [Google Scholar]
- 35.Durnford DG, Price JA, McKim SM, Sarchfield ML. Light-harvesting complex gene expression is controlled by both transcriptional and post-transcriptional mechanisms during photoacclimation in Chlamydomonas reinhardtii. Physiol Plant. 2003;118:193–205. [Google Scholar]
- 36.Khrebtukova I, Spreitzer RJ. Elimination of the Chlamydomonas gene family that encodes the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci U S A. 1996;93:13689–13693. doi: 10.1073/pnas.93.24.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinbothe S, Reinbothe C, Parthier B. Methyl jasmonate–regulated translation of nuclear-encoded chloroplast proteins in barley (Hordeum vulgare L. cv. Salome) J Biol Chem. 1993;268:10606–10611. [PubMed] [Google Scholar]
- 38.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 39.Gibbs S. The route of entry of cytoplasmically synthesized proteins into chloroplasts of algae possessing chloroplast ER. J Cell Sci. 1979;35:253–266. doi: 10.1242/jcs.35.1.253. [DOI] [PubMed] [Google Scholar]
- 40.Chua NH, Blobel G, Siekevitz P, Palade GE. Attachment of chloroplast polysomes to thylakoid membranes in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1973;70:1554–1558. doi: 10.1073/pnas.70.5.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suss KH, Prokhorenko I, Adler K. In situ association of Calvin cycle enzymes, ribulose-1,5-bisphosphate carboxylase/oxygenase activase, ferredoxin-NADP+ reductase, and nitrite reductase with thylakoid and pyrenoid membranes of Chlamydomonas reinhardtii chloroplasts as revealed by immunoelectron microscopy. Plant Physiol. 1995;107:1387–1397. doi: 10.1104/pp.107.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pyhtila B, et al. Signal sequence– and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14:445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Margulies MM, Michaels A. Ribosomes bound to chloroplast membranes in Chlamydomonas reinhardtii. J Cell Biol. 1974;60:65–77. doi: 10.1083/jcb.60.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto T, Burke J, Autz G, Jagendorf AT. Bound ribosomes of pea chloroplast thylakoid membranes: Location and release in vitro by high salt, puromycin, and RNase. Plant Physiol. 1981;67:940–949. doi: 10.1104/pp.67.5.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green-Willms NS, Fox TD, Costanzo MC. Functional interactions between yeast mitochondrial ribosomes and mRNA 5′ untranslated leaders. Mol Cell Biol. 1998;18:1826–1834. doi: 10.1128/mcb.18.4.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt RJ, Myers AM, Gillham NW, Boynton JE. Immunological similarities between specific chloroplast ribosomal proteins from Chlamydomonas reinhardtii and ribosomal proteins from Escherichia coli. Mol Biol Evol. 1984;1:317–334. doi: 10.1093/oxfordjournals.molbev.a040320. [DOI] [PubMed] [Google Scholar]
- 47.Randolph-Anderson BL, Gillham NW, Boynton JE. Electrophoretic and immunological comparisons of chloroplast and prokaryotic ribosomal proteins reveal that certain families of large subunit proteins are evolutionarily conserved. J Mol Evol. 1989;29:68–88. doi: 10.1007/BF02106183. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt RJ, Richardson CB, Gillham NW, Boynton JE. Sites of synthesis of chloroplast ribosomal proteins in Chlamydomonas. J Cell Biol. 1983;96:1451–1463. doi: 10.1083/jcb.96.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleming GH, Boynton JE, Gillham NW. The cytoplasmic ribosomes of Chlamydomonas reinhardtii: Characterization of antibiotic sensitivity and cycloheximide-resistant mutants. Mol Gen Genet. 1987;210:419–428. doi: 10.1007/BF00327192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.