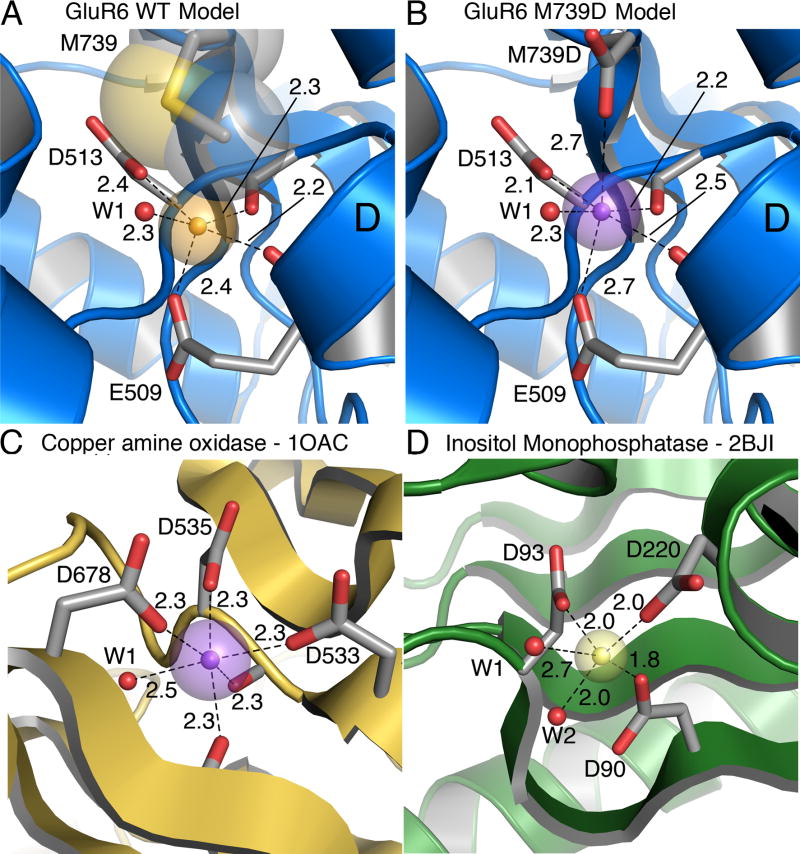
Figure 5. Model divalent ion binding sites.
(A) Structural model of the GluR6 wild-type cation binding site based on the Na-GluR5 complex (3c32), with sodium as a gold sphere. Transparent spheres represent the Shannon radius. The Methionine residue that caps the binding site is shown in CPK representation.
(B) Structural model of the GluR6 M739D mutant with calcium bound (violet sphere). The model is derived from the Na-GluR5 complex (3c32) with the ‘capping’ isoleucine replaced by an aspartic acid. The bound sodium ion was replaced by a calcium ion (violet sphere) placed equidistant between the proximal carboxyl groups of E509 and M739D.
(C) Ca (violet sphere) bound to copper amine oxidase (1OAC) with five protein ligands. The Ca ion is buried but the crown of three aspartic acid side chains that coordinate it is partly solvent exposed. Two backbone carbonyl oxygen atoms and a water molecule (W1) complete the six-fold coordination.
(D) Mg (yellow sphere) bound in the active site of inositol monophosphatase in a substrate-free crystal form (2BJI). The coordination sphere has three carboxylates and two waters (red spheres, W1 and W2). A site is available for third water molecule to give octahedral coordination, although it was not modeled.