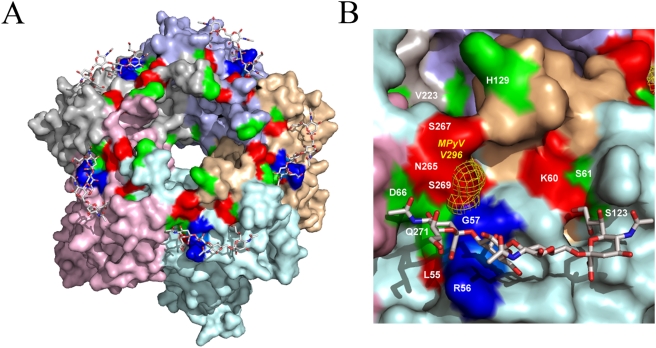
Figure 2. Structural model of JCV VP1/NeuNAc–(α2,3)–Gal–(β1,3)–[(α2,6)-NeuNAc]–Glc-NAc tetrasaccharide complex.
(A) A model of JCV VP1 basic pentamer in complex with NeuNAc–(α2,3)–Gal–(β1,3)–[(α2,6)-NeuNAc]–Glc-NAc tetrasaccharide. Surfaces of five chains of JCV VP1 are shown in different colors. The RG motif essential for binding of core sialic acid is shown in blue. PML-associated mutated residues confirmed by PAML (Table 1) are shown in red (L55, K60, S265, S267, S269). Additional mutations unique to PML-isolated samples (Table S2) are shown in green (S61, D66, S123, H129, V223 and Q271). (B) A close-up view of NeuNAc–(α2,3)–Gal–(β1,3)–[(α2,6)-NeuNAc]–Glc-NAc tetrasaccharide/JCV VP1 complex. The color scheme is as described in panel (A). Location of V296 of MPyV VP1 which is predicted to be equivalent to S269 of JCV VP1 is shown in yellow mesh.