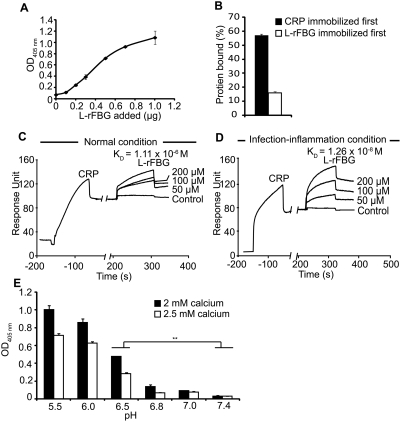
Figure 5. Biochemical characterization of the interaction between CRP and L-rFBG.
(A) Dose-dependent binding of L-rFBG to 0.8 µg of immobilized CRP. The indicated amount of L-rFBG in blocking buffer was added to the immobilized CRP. OD405 nm was read to indicate the binding of L-rFBG to CRP. 0.8 µg of HSA immobilized on the wells instead of CRP was the negative control and readings were subtracted off the negative controls. (B) Effect of the order of immobilization on the binding between CRP and L-rFBG. 0.8 µg of L-rFBG was added to 0.8 µg of immobilized CRP (black bar) and 0.8 µg of CRP was added to 0.8 µg of immobilized L-rFBG (white bar). To compare the data from the two orders of immobilization, the binding proteins (L-rFBG or CRP) directly immobilized on the plate were detected and were taken as 100% binding, to which other readings were normalized against. The affinity between CRP and L-rFBG under normal (C) or infection-inflammation condition (D) was tested by surface plasmon resonance analysis using BIAcore 2000. POPC was immobilized on the HPA chip to expose PC as ligand for CRP. A 50 µl of CRP at 200 nM in the running buffer was injected. Fifty µl aliquots of L-rFBG at the indicated concentrations were injected afterwards. The negative controls involved injection of buffers (without L-rFBG) over the PC-HPA chip immobilized with CRP. Data were analyzed using BIAevaluation 3.2 software. (E) Binding of L-rFBG to immobilized CRP at pH ranging from 5.5 to 7.4, in the presence of 2 mM calcium (black bars) or 2.5 mM calcium (white bars). HSA instead of CRP immobilized on the plates was the negative control and readings were subtracted off the negative controls.