Abstract
Background
The Phosphatidylinositol 3′-kinase is a key regulator in various cancer-associated signal transduction pathways. Genetic alterations of its catalytic subunit alpha, PIK3CA, have been identified in ovarian cancer. Our in vivo data suggests that PIK3CA activation is one of the early genetic events in ovarian cancer. However, its role in malignant transformation of ovarian surface epithelium (OSE) is largely unclear.
Methodology/Principal Findings
Using the Müllerian inhibiting substance type II receptor (MISIIR) promoter, we generated transgenic mice that expressed activated PIK3CA in the Müllerian epithelium. Overexpression of PIK3CA in OSE induced remarkable hyperplasia, but was not able to malignantly transform OSE in vivo. The consistent result was also observed in primary cultured OSEs. Although enforced expression of PIK3CA could not induce OSE anchorage-independent growth, it significantly increased anchorage-independent growth of OSE transformed by mutant K-ras.
Conclusions/Significance
While PIK3CA activation may not be able to initiate OSE transformation, we conclude that activation of PIK3CA may be an important molecular event contributing to the maintenance of OSE transformation initiated by oncogenes such as K-ras.
Introduction
Epithelial ovarian cancer continues to be the leading cause of death among gynecological malignancies [1], [2], [3]. The lack of effective methods for prevention, early detection and treatment recurrent ovarian tumors creates a pressing need to understand its pathogenesis and identify molecular targets for therapy. Cancer is a disease involving multistep dynamic changes in the genome [4]. However, the oncogenic events and their cooperation that promote malignant transformation in ovarian carcinoma remain largely unknown [2], [3], [5], [6], [7], [8], [9], [10].
Phosphatidylinositol-3′ kinase (PI-3 kinase) is an intracellular signal transducer with lipid substrate specificity implicated in a wide range of cancer-associated signaling pathways including tumor cell metabolism, survival and proliferation [11], [12], [13], [14], [15], [16], [17], [18], [19]. It is recruited and activated by multiple receptor tyrosine kinases and generates second messengers via phosphorylation of membrane inositol lipids at the D3 position [11], [12], [13], [14]. PI-3 kinase was first recognized as a putative oncogene because of its ability to bind polyoma middle T antigen [20], [21]. Molecular cloning of PI-3 kinases revealed a large and complex family that contains three classes with multiple subunits and isoforms [11], [12], [13], [14]. The PIK3CA gene encodes the catalytic subunit p110-alpha, one of the three catalytic subunit proteins of the class IA PI-3 kinases [11], [12], [13], [14]. PIK3CA was identified as an avian retrovirus-encoded oncogene that can transform chicken embryo fibroblasts [22]. Numerous recent studies indicate that PIK3CA and downstream pathways are frequently targeted by genomic amplification [23], mutation [24] or overexpression [25] in solid tumors including ovarian cancer [23], [25], [26], [27], [28], [29], [30], [31], [32]. Previous studies on the function of PIK3CA in ovarian cancers have been predominantly focused on the maintenance and survival of late-stage of ovarian carcinoma. The function of PIK3CA in malignant transformation of the ovarian surface epithelium (OSE) remains unexplored. Here we generated transgenic mice expressing constitutively activated (myristoilated) PIK3CA in the Müllerian epithelium of the female genital tract to investigate the effect of PIK3CA overexpression in the OSE.
Results
PIK3CA overexpression was an early genetic event during ovarian tumorigenesis
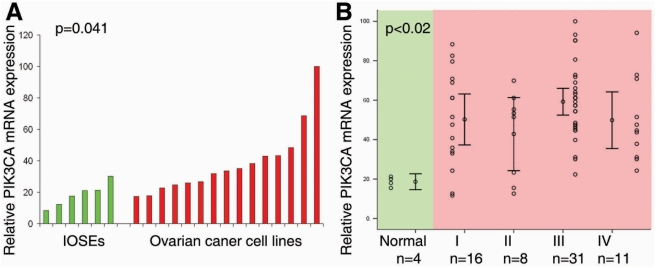
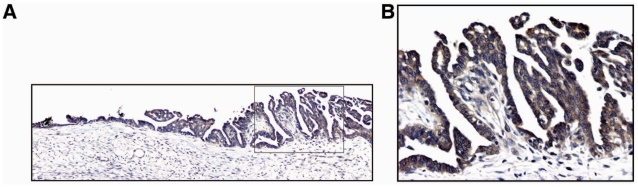
In many human tumors, including epithelial ovarian cancer [23], [25], [26], [27], [28], [29], [30], PIK3CA activation is a critical oncogenic event and can be mediated by multiple genetic/genomic alterations such as gene copy number amplification [23], gain of functional mutations [24], and transcriptional up-regulation [33], [34], [35]. However, the function of PIK3CA activation during the process of malignant transformation of OSE is still not well understood. Our previous studies indicate that mRNA expression of PIK3CA is significantly up-regulated in the early-stage of ovarian cancer development, strongly suggesting that PIK3CA might be involved in OSE transformation [25]. To confirm our previous finding, we first compared expression levels of PIK3CA mRNA in established epithelial ovarian cancer cell lines (n = 15) with primary cultures of immortalized human ovarian surface epitheliums (IOSEs, n = 6). Consistently, we found that mRNA levels in epithelial tumor cell lines were significantly higher than in IOSEs (p = 0.041, Figure 1A). Next, we examined mRNA expression of PIK3CA in microdissected normal human ovarian epithelium (n = 4) as well as epithelial ovarian cancer specimens including FIGO stages I (n = 16), II (n = 8), III (n = 31) and IV (n = 11). We found that mRNA expression level of PIK3CA was significantly upregulated in ovarian cancer specimens compared to normal control ovarian epithelium (p<0.02), and there was no further significant increase after malignant transformation among different stages of ovarian cancer (p>0.05, Figure 1B), which is consistent with our previous observation [25]. To further confirm PIK3CA is indeed expressed in early-stage ovarian cancer, we also examined the protein product of PIK3CA gene, p110α, by immunohistochemical staining in early malignant transformed human ovarian surface epithelium. We found that p110α was highly detectable in the early malignant transformed human ovarian surface epithelium (Figure 2A and B). Taken together, these results indicate that PIK3CA overexpression is in fact an early genetic event during ovarian oncogenesis, thus suggesting that PIK3CA activation might be causally involved in this process.
Figure 1. PIK3CA overexpression was an early genetic event during ovarian tumorigenesis.
A. PIK3CA mRNA expression was significantly up-regulated in the established epithelial ovarian cancer cell lines (n = 15) compared with primary cultured ovarian surface epithelial cells (n = 6, p = 0.041). mRNA expression was measured by real-time RT-PCR. B. mRNA expression level of PIK3CA was significantly upregulated in ovarian cancer specimens compared with normal control ovarian epithelium. Normal ovarian epithelium was isolated by laser-capture microdissection. There was no further significant increase of PIK3CA mRNA expression after malignant transformation among different stages of ovarian cancer.
Figure 2. High p110α expression was detected in early malignant transformed human ovarian surface epithelium.
p110α expression was detected by immunohistochemical staining. A. High p110α expression was detected in early malignant transformed human ovarian surface epithelium (100X). B. High magnification of A (400X).
Generation of transgenic mice expressing activated PIK3CA in the Müllerian epithelium
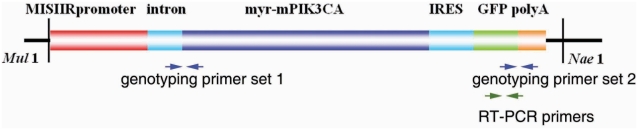
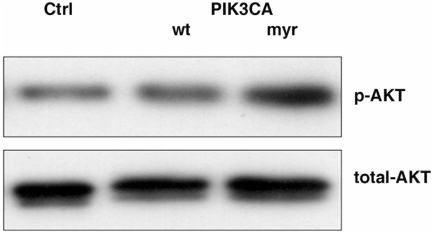
To investigate the role of PIK3CA in malignant transformation of the OSE in vivo, we generated transgenic mice in which activated PIK3CA was specifically overexpressed in the Müllerian epithelium of the female reproductive tract including OSE. In this model, we used the Müllerian epithelium specific promoter, Müllerian inhibiting substance type II receptor promoter (MISIIR) [36], [37], [38], [39], to drive expression of the murine PIK3CA (Figure 3). In the male animal, Müllerian inhibiting substance (MIS) is secreted from Sertoli cells of the developing testes and stimulates the regression of the Mullerian duct. Testosterone is also secreted from the developing testes and induces the differentiation of the Wolffian duct into the secondary structures of the male reproductive tract. In the absence of MIS in the developing female embryo, the Mullerian duct differentiates into the secondary structures of the female reproductive tract [36], [37], [38], [39]. Expression of the MISIIR has been reported to be restricted to mesenchymal cells surrounding the Mullerian duct during embryogenesis, tubular and follicular structures of fetal gonads, Sertoli and Leydig cells of adult testis, and granulosa cells of adult ovary [36], [37], [38], [39]. Above information provides a possible strategy to develop a transgenic model of ovarian carcinoma [39]. Using this promoter, Connolly et al. have successfully developed the very first ovarian cancer transgenic models that develop ovarian carcinomas with metastatic spread to peritoneal organs [39]. In addition, increasing evidence indicates that PIK3CA is activated in a large percentage of human ovarian cancer patients [25], [27], [35]. Therefore, we generated the activating mutation by addition of the avian src myristoylation sequence (MGSSKSKPK) at the N-terminus of the wild type of murine PIK3CA to constitutively activate PI3-kinase pathway in vivo. To demonstrate that our transgenic construct was able to constitutively activate PI3-kinase pathway, we transient transfected myr-PIK3CA, wt-PIK3CA and control vectors to ovarian cancer cell line 2008. 48 hrs of post-transfection, the transfected cells were cultured in low serum overnight. Protein and total RNA were isolated from cells. Real-time RT-PCR demonstrated that cells from wt-PIK3CA and myr-PIK3CA transfections were expressed similar levels of PIK3CA mRNA, which was about 11.5-fold higher comparing to cells from control vector transfection. Total and phosphate AKT, the downstream molecule of PI3-kinase pathway, were examined by western blot. Figure 4 showed that myr-PIK3CA was able to constitutively activate AKT in low serum condition (1%) compared with wt-PIK3CA and control transfection.
Figure 3. The illustration of the construct that was used to generate transgenic mouse.
The arrows show two sets of the genotyping primers and one set of the RT-PCR primers.
Figure 4. myr-PKI3CA was able to active PI3-kinase pathway.
myr-PIK3CA, wt-PIK3CA and control vectors were transient transfected to ovarian cancer cell line 2008. 48 hrs of post-transfection, transfected cells were cultured in 1% serum over night. Total and phosphate AKT were detected by western blot.
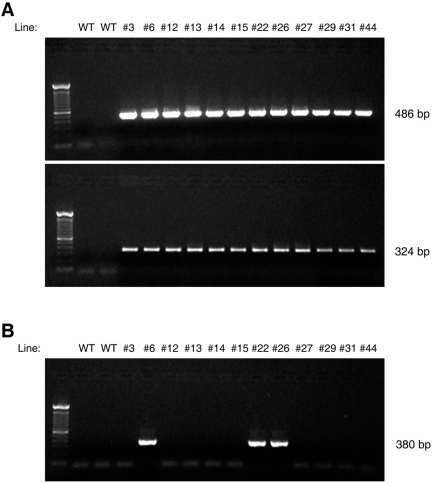
Twelve founder lines of MISIIR-PIK3CA transgenic (PIK3CA-Tg) mice were generated, in which the genomic integration of whole MISIIRpr-myrPIK3CA-IRES-GFP sequence was confirmed by two sets of genotyping primers (Figure 5A). Expression of the transgene mRNA (eGFP) was examined by RT-PCR in the samples from the female whole ovary of the genotype-positive mice. In three of the transgenic lines, the eGFP mRNA transcript was able to be detected in vivo (Figure 5B). Three founder PIK3CA-Tg lines (#6, #22 and #26), three control lines (genotype positive, eGFP mRNA negative #3, #12 and #44, Figure 5B) and wild type mice (genotype negative mice) were used for further studies.
Figure 5. Generation of transgenic mice expressing activated PIK3CA in the Müllerian epithelium.
A. A total of 12 founder lines were generated. Upper panel shows the genotyping by PCR using the first set of the genotyping primers. Lower panel shows the genotyping by PCR using the second set of the genotyping primers. B. The transgene (eGFP) was able to be detected in 3 founder lines by RT-PCR.
Overexpression of activated PIK3CA in OSE resulted in hyperplasia but not tumor formation
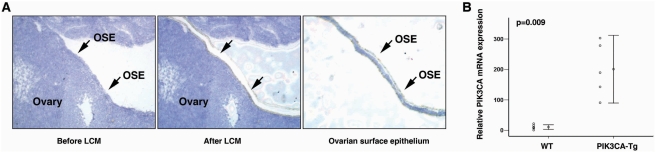
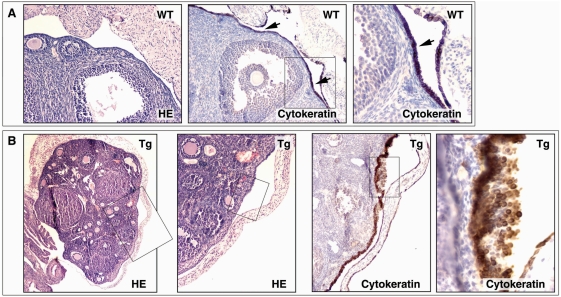
To confirm overexpression of PIK3CA in the OSEs of transgenic mouse, we microdissected the OSE cells from PIK3CA-Tg and WT mice using laser capture microdissetion (LCM, Figure 6A). Total RNA was isolated, and PIK3CA mRNA expression was measured by real-time RT-PCR. We found that PIK3CA mRNA expression level was 18.5-fold higher in OSEs from PIK3CA-Tg mice (200.93 ±89.47 relative expression unit) than from WT mice (10.87 ± 7.28 relative expression unit, p = 0.009, Figure 6B). In addition, by immunohistochemistry (using two different antibodies), we confirmed that p110α was highly expressed in the OSE of the PIK3CA-Tg mice (data not shown). Then, we followed up the ovarian tumor development in PIK3CA-Tg mice. We found that expression of the activated PIK3CA resulted in hyperplasia in mouse OSE and a paucity of follicles in four month-old female mice. Female transgenic PIK3CA-Tg mice exhibited subfertility. Figure 7A shows the typical OSE in 5-month control female mice. The Cytokeratin (an epithelial marker) positive OSE was observed on the surface of ovary as a monolayer. In the PIK3CA-Tg mice, hyperplasia was found in the OSE, and the epithelial origin of the lesions was confirmed by Cytokeratin staining (Figure 7B). Hyperplasia was found in more than 50% of the PIK3CA-Tg mice after four months post-birth, and in 100% of the PIK3CA-Tg mice after ten months post-birth. There were no invaginations or papillary structures observed in the OSE of PIK3CA-Tg mice. In the control mice, no significant hyperplasia in OSE was observed even after 12 months of post-birth. These results suggest that expression of activated PIK3CA in OSE induced hyperplasia of the OSE. We monitored for ovarian tumor development in both the transgenic and control mouse lines. A total of 218 female PIK3CA-Tg mice (#6: n = 94; #22: n = 44 and #26: n = 80) were evaluated (at least 30 mice of each line were followed for more than 18 months) and no epithelial ovarian tumors were observed. There was no difference in life span between the transgenic mice and control lines. This finding indicates that expression of activated PIK3CA in OSE causes hyperplasia in vivo but not tumor formation.
Figure 6. PIK3CA was overexpressed in OSE of PIK3CA-Tg mouse.
A. Ovarian surface epithelial cells were microdissected by laser capture microdissection technology. B. PIK3CA mRNA expression in OSEs from WT or PIK3CA-Tg OSEs was analyzed by real-time RT-PCR. The primers were able to detected both wt PIK3CA cDNA and cDNA from transgene expression (myr-PIK3CA).
Figure 7. Overexpression of activated PIK3CA in OSE resulted in hyperplasia but not tumor formation.
A. The ovarian surface epithelium in wild type mouse. B. The ovarian surface epithelium in PIK3CA-Tg mouse. The epithelium origin was confirmed by cytokeratin staining.
PIK3CA contributed to K-ras initiated transformation in vitro
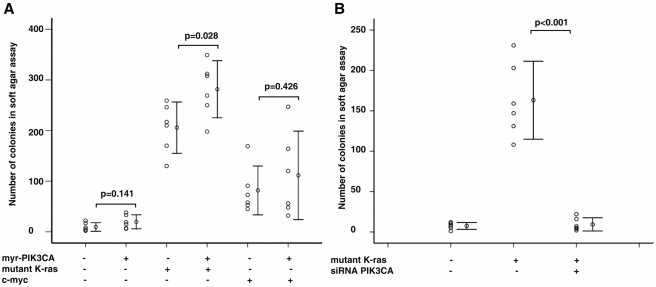
To further confirm the above in vivo observation, we next tested the role of PIK3CA in OSE transformation in vitro. Primary murine OSE cells (MOSEs) were isolated from ovaries of female mice and cultured short-term (only early passage, <10 passages, cells were used in this study) [40]. Soft agar assay was used to evaluate the potential contribution of PIK3CA in transformation of OSE [41]. Because PI3-kinase/PTEN-ras [8] and PI3-kinase/AKT-myc [5] pathways have been reported involving in OSE transformation, we chose ras and c-myc in our following studies. Myr-PIK3CA (pUSEamp-myr-mPIK3CA, under CMV promoter, G418), K-rasv12 and c-myc (both in pBabe-puro vector, puromycin) expression vectors were transfected to MOSE cells. The pooled stable expression cells were generated by short-term antibiotic selection (10 days for G418 or 3 days for puromycin). The transgene expression was further confirmed by RT-PCR. We found that overexpression of PIK3CA alone did not significantly increase anchorage-independent growth of OSEs (Figure 8A). In contrast, introduce of either mutant K-ras of c-myc resulted in increased colony formation of OSEs (Figure 8A). This result indicates that unlike mutant K-ras or c-myc, PIK3CA is not able to cause complete transformation of OSE in vitro. Next, we tested the combination of PIK3CA with either of the two other oncogenes by co-transfection. The myr-PIK3CA was transfected first. After 10 days G418 selection, K-ras or c-myc was transfected subsequently and selected again by puromycin for 3 days. Interestingly, we found that combining PIK3CA and mutant K-ras significantly increased anchorage-independent growth of cultured OSE cells compared with mutant K-ras alone (Figure 8A). However, PIK3CA did not significantly increase colony numbers of OSE cells transformed by c-myc (Figure 8A). This finding suggests that early activation of PIK3CA might promote transformation of OSE cells in certain cellular and molecular contexts, e.g. in the presence of K-ras mutation. To further confirm that PIK3CA can play a role in transformation induced by mutant K-ras, we blocked endogenous PIK3CA expression in cultured OSE cells transfected with mutant K-ras by RNA interference using small interfering RNAs (siRNA). The efficiency of the siRNA targeting mPIK3CA was confirmed by real-time RT-PCR (mRNA expression of mPIK3CA was knocked down to ∼30% in the siRNA treated cells compared with control cells, data not shown). The specificity of the siRNA was also examined by measuring endogenous mPIK3CB expression (there was no significantly difference of PIK3CB mRNA expression between siRNA treated and control cells, data not shown). We found that in the absence of PIK3CA, the transformed OSEs could not grow in soft agar (Figure 8B), which suggests that the growth of K-ras transfected OSE cells require PIK3CA expression. However, it is still unclear whether PIK3CA contributes to K-ras initiated transformation in vivo. Generation of “bigenic” mouse expressing both activated PIK3CA and mutant Ras specifically in murine OSE using MISIIR promoter is still a technical challenge. Because mutant Ras is able to fully transform epithelium in vivo [42], MISIIR-driven mutant Ras will induce very rapidly reproductive system carcinoma in both female and male animals, and these transgenic mice will lose reproductive ability. Therefore, it will be difficult to generate “bigenic” animals by crossing MISIIR-driven mutant Ras and MISIIR-driven myr-PIK3CA mice. We believe that a novel transgenic strategy based on the Cre-loxp conditional expression and intrabursal administration of adenovirus [6], [8], [9] may allow us to further test PIK3CA and Ras cooperation in vivo in the future.
Figure 8. PIK3CA contributed to K-ras initiated transformation in vitro.
A and B. Summary of colony numbers of the different combination of oncogenes in soft agar assay.
Discussion
In agreement with our findings, several groups have demonstrated that activation of PI3-kinase plays a crucial role in epithelial cell transformation. For example, in human mammary epithelial cells (HMECs), activation of the PI3-kinase pathway in the presence of increased expression of c-myc, can functional replace small T antigen and result in anchorage-independent growth [43]. In p53 null murine OSE cells, enforced expression of the PI3-kinase downstream mediator, Akt, in cooperation with mutant K-ras, induces the OSE cell transformation [5]. Moreover, two independent laboratories have recently reported that conditional Pten deletion combined with mutant K-ras [8] or deregulated Wnt/Catenin pathway [9] is able to induce endometrioid ovarian tumors in mice in vivo. In all of these studies, only activation of wild type PIK3CA alone seems insufficient to initiate epithelial transformation, consistent with the results of our study. Although mutant PIK3CA has been reported to be sufficient to transform normal cells both in vitro [44], [45], [46] and in vivo [46], [47], PIK3CA mutation exhibits a relatively low frequency in ovarian cancer (4.8–12%) [28], [29], [30], [32] as compared to other cancer types. In addition, PIK3CA mutation is rare in borderline [30] or early-stage ovarian tumors (our unpublished observation). Therefore, PIK3CA mutation might not be commonly involved in the transformation of the OSE. However, PIK3CA amplification is one of most common genetic alterations in ovarian cancer (23.6–40%) [23], [26], [27], [31] and more importantly, an increase in copy number at chromosomal locus 3q26-qter, which harbors the PIK3CA gene, has been observed in 40% of early-stage ovarian cancers [26], suggesting that PIK3CA amplification might be one of the critical events in OSE transformation. In this study, we tested overexpression of activated wild type PIK3CA in OSE in vivo and in vitro, since it might more closely resemble natural OSE transformation during human ovarian cancer development. Taken together, we conclude that PIK3CA activation is one of the early molecular events during OSE transformation, and activation of PIK3CA contributes to tumorigenesis in certain cellular and molecular contexts. However, PIK3CA activation might not be the initial event in OSE transformation, and may require cooperation with other oncogenic events such as K-ras mutation to maintain transformed OSE growth.
Materials and Methods
Patients and Specimens
The specimens used in this study were collected at the University of Pennsylvania and the University of Turin, Italy. All tumors were from primary sites, and were immediately snap-frozen and stored at −80°C. Ethical approval for this work was granted by institution's Institutional Review Boards (IRBs) of the University of Pennsylvania and the University of Turin. Tissues were obtained after informed written consent from patients involved under a general tissue collection protocol approved by the IRBs.
Cell lines and Cell Culture
A total of 15 ovarian cell lines were used in this study. All cancer cell lines were cultured in DMEM medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Invitrogen). Six independent immortalized human ovarian surface epithelial cells (IOSEs, generously provided by Drs. Auersperg and Birrer) were cultured in 1∶1 media 199: MCDB 105 (Sigma, St. Louis, MO) supplemented with 15% FBS. Murine ovarian surface epithelial cells (MOSEs) were isolated and cultured as previously reported [40].
Plasmid Construction
pMISIIRprom-TOPO, which contains the murine Müllerian inhibitory substance type II receptor (MISIIR) gene 5′ regulatory region, was constructed by PCR amplification from mouse genomic DNA [39]. pUSEamp-myr-mPIK3CA, which contains murine PIK3CA under the control of CMV promoter, was ordered from UPSTATE (UPSTATE, Lake Placid, NY). The activating mutation was generated by addition of the avian src myristoylation sequence (MGSSKSKPK) at the N-terminus. pCI-neo, which contains a chimeric intron, was ordered from Promega (Promega, Madison, WI). pMigR, which contains an internal ribosome entry site (IRES) and downstream enhanced green fluorescent protein (eGFP), was generously provided by Dr. Pear. We tagged myr-PIK3CA in pUSEamp with eGFP, preceded by an IRES. The IRES and eGFP were derived from pMigR. The XhoI-SalI-blunted fragment containing the IRES and eGFP from plasmid pMigR was inserted into the XhoI-ApaI-blunted plasmid pUSEamp-myr-PIK3CA to construct plasmid pUSEamp-myr-PIK3CA-IRES-GFP. The SacI-XhoI linearized fragment containing the chimeric intron from plasmid PCI-neo was inserted into the EcoRV-XhoI linearized plasmid pMISIIRprom-TOPO to construct plasmid pMISIIRprom-Intron. The MluI-ScaI linearized fragment containing the MISIIR promoter and the intron from plasmid pMISIIRprom-Intron was swapped into plasmid pUSEamp-myr-PIK3CA-IRES-GFP to make plasmid pMISIIRprom-Intron-myr-PIK3CA-IRES-GFP, in which the CMV promoter has been replaced by the MISIIR promoter and the intron upstream of the PIK3CA ORF, as well as the IRES, eGFP reporter and hGH polyA downstream (for the detailed information for construction also see Figure S1). This construct was used to generate a transgenic mouse expressing myr-PIK3CA in the OSE.
Protein Isolation and Western blot
Cultured cells were lysed in 200 μl of lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 1% Triton X-100. Protein was separated by 10% SDS-PAGE under denaturing conditions and transferred to nitrocellulose membrane. Membranes were incubated with an anti-total AKT or anti-pAKT antibodies (1∶1,000, Cell Signaling Technology, Danvers, MA), followed by incubation in anti-rabbit secondary antibody conjugated with horseradish peroxidase (1∶10,000; Cell Signaling Technology). Immunoreactive proteins were visualized using enhanced chemiluminescence detection system (Amersham Biosciences, Piscataway, NJ).
Transgenic Mice
The animal study protocol was reviewed and approved by the institutional animal care and use committee (IACUC) of the University of Pennsylvania. Transgenic mice were generated by the University of Pennsylvania's Transgenic & Chimeric Mouse Facility. The linearized transgene DNA fragment (MISIIR-myr-PIK3CA-IRES-GFP) was injected by microinjection into pronuclei of day-0.5 embryos of the first generation of a hybrid genetic background of C57BL/6 and C3H (B6C3F1) mice. Injected embryos were implanted into the oviducts of day-0.5 pseudo pregnant female Swiss Webster mice.
Genotyping
Tails from the resulting pups were clipped 3 weeks after birth for genotype analysis. Genomic DNA was isolated by the DNeasy Tissue kit (QIAGEN, Valencia, CA). Presence of the transgene was confirmed by PCR amplification of a 486-bp fragment (Frag1) of the intron-myr-PIK3CA as well as a 324–bp fragment (Frag2) of the eGFP. Specific primers for eGFP were as follows: Frag1 forward primer: 5′-AGG CAC TGG GCA GGT AAG TAT, Frag1 reverse primer: 5′-CAT GTT TGA TGG TGA CGA GTG; Frag2 forward primer: 5′-CGA CAA CCA CTA CCT GAG CA, Frag2 reverse primer: 5′-TTA GGA AAG GAC AGT GGG AGT G. Genomic DNA (2 μl) was amplified in 25 μl of the PCR reaction containing 200 μmol/L each dNTP, 20 pmol of each primer, the standard buffer supplemented with 1.5 U Taq polymerase (Roche, Indianapolis, IN), and 1.5 mmol/L MgCl2. After initial denaturation at 94°C for 4 minutes, 30 cycles of PCR were performed with denaturation at 94°C for 15 seconds, annealing at 55°C for 20 seconds, and extension at 72°C for 45 seconds. The last extension was at 72°C for 7 minutes.
RNA Isolation and Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated from 100 to 500 mg of fresh tissue or 1×106 cultured cells with TRIzol reagent (Invitrogen). Total RNA was reverse-transcribed using the Superscript first-strand synthesis kit for RT-PCR (Invitrogen). Reverse-transcribed cDNA was amplified in 25 μl of the PCR reaction containing 200 μmol/L each dNTP, 20 pmol of each primer, the standard buffer supplemented with 1.5 U Taq polymerase (Roche), and 1.5 mmol/L MgCl2. After initial denaturation at 94°C for 4 minutes, 35 cycles of PCR were performed with denaturation at 94°C for 15 seconds, annealing at 55°C for 20 seconds, and extension at 72°C for 45 seconds. The last extension was at 72°C for 7 minutes. Specific primers for GFP were as follows: eGFP forward primer: 5′-AGC TGA CCC TGA AGT TCA TCT G, GFP reverse primer: 5′-GAT CTT GAA GTT CAC CTT GAT GC.
Real-time RT-PCR
cDNA was quantified by real-time RT-PCR on the ABI Prism 7900 Sequence Detection System (Applied Biosystems, Foster City, CA). PCR was performed using Sybr Green PCR Core reagents (Applied Biosystems). PCR amplification of the housekeeping genes GAPDH was performed for each sample as a control for sample loading and to allow normalization among samples. A standard curve was constructed containing the PIK3CA cDNA and amplified by the real-time PCR. Each sample was run in duplicate and each PCR experiment included two non-template control wells.
Laser capture microdissection (LCM)
LCM was performed as described by our previous study [48]. Briefly, cryosections (10 μm) from human or mouse ovaries were mounted on a polyethylene foil slide (SL Microtest, Jena, Germany). After rapid hematoxylin staining, sections were subjected to LCM utilizing the μCUT Laser-MicroBeam System (SL Microtest) with a fine ultraviolet laser, which enables the contact-free isolation of single cells or groups of cells. The microdissected cells were catapulted into the lid of a 0.5-ml reaction tube containing RNA isolation buffer. RNA was isolated by TRIzol reagent.
Immunostaining
Immunohistochemical staining was performed using the avidin-biotin-peroxidase method [49]. Sections were pretreated with 0.03% H2O2 for 20 minutes to block endogenous peroxidase activity and incubated in matched normal sera (Vector Laboratories, Burlingame, CA). The following primary antibodies were used in this study: Cytokeratin (Hybridoma Bank, Iowa City, IA, 1∶200) rabbit anti-p110α (Cell Signaling, Danvers, MA 1∶200) and mouse anti-human p110a (Pharmigen, CA, 1∶200). Mouse on Mouse (M.O.M.) kit (Vector) was used for monoclonal antibody. The Vectastain ABC kit was applied as described by the manufacturer (Vector Laboratories). Sections were counterstained with Gill's hematoxylin (Vector Laboratories). Images were acquired through a Cool SNAP Pro color digital camera (Media Cybernetics).
RNA interference/Transfection of Synthetic siRNA
Synthetic SMARTpool siRNAs targeting mouse PIK3CA (Dharmacon, Chicago, IL) or appropriate siCONTROL non-targeting siRNAs (Dharmacon) were transfected into cultured cells. Transfection was performed using LipofectamineTM2000 (Invitrogen) following the manufacturer's instructions. Forty-eight hrs post transfection, total RNA was extracted to examine the PIK3CA expression by real-time RT-PCR.
In vitro Cell Transformation Assay
A soft agar assay was performed using Cell Transformation Detection Assay Kit (Chemicon, Temecula, CA) following the manufacturer's instructions [41].
Statistics
Statistical analysis was performed using the SPSS statistics software package (SPSS, Chicago, IL). All results were expressed as mean±SD, and p<0.05 was used for significance.
Supporting Information
(0.05 MB PDF)
Acknowledgments
We thank Dr. Jean Richa (University of Pennsylvania) and the University of Pennsylvania Transgenic & Chimeric Mouse Facility for the transgenic mouse generation; Drs. Steven Johnson and Kang-Shen Yao (University of Pennsylvania) for the human ovarian cancer cells; Dr. Michael J. Birrer (NCI) for HOSE cells; Dr. Nelly Auersperg (University of British Columbia) for HOSE cells and access to the Canadian Ovarian Tissue Bank; Dr. Pear (University of Pennsylvania) for pMigR vector; and Saundra Ehrlich for editing assistance on this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Ovarian Cancer Research Found (LZ), NCI Ovarian Cancer SPORE P50-CA83638 (Career Development Award, LZ), American Cancer Society IRG-78-002-30 (LZ), and Mary Kay Ash Charitable Foundation (LZ). DK was partly supported by the Italian Association for Cancer Research (AIRC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, et al. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Auersperg N, Wong AS, Choi KC, Kang SK, Leung PC. Ovarian surface epithelium: biology, endocrinology, and pathology. Endocr Rev. 2001;22:255–288. doi: 10.1210/edrv.22.2.0422. [DOI] [PubMed] [Google Scholar]
- 3.Landen CN, Jr, Birrer MJ, Sood AK. Early events in the pathogenesis of epithelial ovarian cancer. J Clin Oncol. 2008;26:995–1005. doi: 10.1200/JCO.2006.07.9970. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Orsulic S, Li Y, Soslow RA, Vitale-Cross LA, Gutkind JS, et al. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell. 2002;1:53–62. doi: 10.1016/s1535-6108(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flesken-Nikitin A, Choi KC, Eng JP, Shmidt EN, Nikitin AY. Induction of carcinogenesis by concurrent inactivation of p53 and Rb1 in the mouse ovarian surface epithelium. Cancer Res. 2003;63:3459–3463. [PubMed] [Google Scholar]
- 7.Liu J, Yang G, Thompson-Lanza JA, Glassman A, Hayes K, et al. A genetically defined model for human ovarian cancer. Cancer Res. 2004;64:1655–1663. doi: 10.1158/0008-5472.can-03-3380. [DOI] [PubMed] [Google Scholar]
- 8.Dinulescu DM, Ince TA, Quade BJ, Shafer SA, Crowley D, et al. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11:63–70. doi: 10.1038/nm1173. [DOI] [PubMed] [Google Scholar]
- 9.Wu R, Hendrix-Lucas N, Kuick R, Zhai Y, Schwartz DR, et al. Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/beta-catenin and PI3K/Pten signaling pathways. Cancer Cell. 2007;11:321–333. doi: 10.1016/j.ccr.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 10.Chodankar R, Kwang S, Sangiorgi F, Hong H, Yen HY, et al. Cell-nonautonomous induction of ovarian and uterine serous cystadenomas in mice lacking a functional Brca1 in ovarian granulosa cells. Curr Biol. 2005;15:561–565. doi: 10.1016/j.cub.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 11.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 12.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 13.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 14.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 15.Jia S, Liu Z, Zhang S, Liu P, Zhang L, et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008. [DOI] [PMC free article] [PubMed]
- 16.Parsons R. Phosphatidylinositol 3-kinase inhibitors are a triple threat to ovarian cancer. Clin Cancer Res. 2005;11:7965–7966. doi: 10.1158/1078-0432.CCR-05-1681. [DOI] [PubMed] [Google Scholar]
- 17.Hu L, Hofmann J, Lu Y, Mills GB, Jaffe RB. Inhibition of phosphatidylinositol 3′-kinase increases efficacy of paclitaxel in in vitro and in vivo ovarian cancer models. Cancer Res. 2002;62:1087–1092. [PubMed] [Google Scholar]
- 18.Hu L, Hofmann J, Jaffe RB. Phosphatidylinositol 3-kinase mediates angiogenesis and vascular permeability associated with ovarian carcinoma. Clin Cancer Res. 2005;11:8208–8212. doi: 10.1158/1078-0432.CCR-05-0206. [DOI] [PubMed] [Google Scholar]
- 19.Saal LH, Johansson P, Holm K, Gruvberger-Saal SK, She QB, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimoto Y, Whitman M, Cantley LC, Erikson RL. Evidence that the Rous sarcoma virus transforming gene product phosphorylates phosphatidylinositol and diacylglycerol. Proc Natl Acad Sci U S A. 1984;81:2117–2121. doi: 10.1073/pnas.81.7.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 22.Chang HW, Aoki M, Fruman D, Auger KR, Bellacosa A, et al. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276:1848–1850. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 23.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 24.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Yang N, Katsaros D, Huang W, Park JW, et al. The oncogene phosphatidylinositol 3′-kinase catalytic subunit alpha promotes angiogenesis via vascular endothelial growth factor in ovarian carcinoma. Cancer Res. 2003;63:4225–4231. [PubMed] [Google Scholar]
- 26.Suzuki S, Moore DH, 2nd, Ginzinger DG, Godfrey TE, Barclay J, et al. An approach to analysis of large-scale correlations between genome changes and clinical endpoints in ovarian cancer. Cancer Res. 2000;60:5382–5385. [PubMed] [Google Scholar]
- 27.Zhang L, Huang J, Yang N, Greshock J, Liang S, et al. Integrative Genomic Analysis of Phosphatidylinositol 3′-Kinase Family Identifies PIK3R3 as a Potential Therapeutic Target in Epithelial Ovarian Cancer. Clin Cancer Res. 2007;13:5314–5321. doi: 10.1158/1078-0432.CCR-06-2660. [DOI] [PubMed] [Google Scholar]
- 28.Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 29.Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama K, Nakayama N, Kurman RJ, Cope L, Pohl G, et al. Sequence mutations and amplification of PIK3CA and AKT2 genes in purified ovarian serous neoplasms. Cancer Biol Ther. 2006;5:779–785. doi: 10.4161/cbt.5.7.2751. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama K, Nakayama N, Jinawath N, Salani R, Kurman RJ, et al. Amplicon profiles in ovarian serous carcinomas. Int J Cancer. 2007;120:2613–2617. doi: 10.1002/ijc.22609. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, Helland A, Holm R, Kristensen GB, Borresen-Dale AL. PIK3CA mutations in advanced ovarian carcinomas. Hum Mutat. 2005;25:322. doi: 10.1002/humu.9316. [DOI] [PubMed] [Google Scholar]
- 33.Singh B, Reddy PG, Goberdhan A, Walsh C, Dao S, et al. p53 regulates cell survival by inhibiting PIK3CA in squamous cell carcinomas. Genes Dev. 2002;16:984–993. doi: 10.1101/gad.973602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Astanehe A, Arenillas D, Wasserman WW, Leung PC, Dunn SE, et al. Mechanisms underlying p53 regulation of PIK3CA transcription in ovarian surface epithelium and in ovarian cancer. J Cell Sci. 2008;121:664–674. doi: 10.1242/jcs.013029. [DOI] [PubMed] [Google Scholar]
- 35.Yang N, Huang J, Greshock J, Liang S, Barchetti A, et al. Transcriptional regulation of PIK3CA oncogene by NF-kappaB in ovarian cancer microenvironment. PLoS ONE. 2008;3:e1758. doi: 10.1371/journal.pone.0001758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Josso N, di Clemente N, Gouedard L. Anti-Mullerian hormone and its receptors. Mol Cell Endocrinol. 2001;179:25–32. doi: 10.1016/s0303-7207(01)00467-1. [DOI] [PubMed] [Google Scholar]
- 37.Teixeira J, Maheswaran S, Donahoe PK. Mullerian inhibiting substance: an instructive developmental hormone with diagnostic and possible therapeutic applications. Endocr Rev. 2001;22:657–674. doi: 10.1210/edrv.22.5.0445. [DOI] [PubMed] [Google Scholar]
- 38.MacLaughlin DT, Donahoe PK. Sex determination and differentiation. N Engl J Med. 2004;350:367–378. doi: 10.1056/NEJMra022784. [DOI] [PubMed] [Google Scholar]
- 39.Connolly DC, Bao R, Nikitin AY, Stephens KC, Poole TW, et al. Female mice chimeric for expression of the simian virus 40 TAg under control of the MISIIR promoter develop epithelial ovarian cancer. Cancer Res. 2003;63:1389–1397. [PubMed] [Google Scholar]
- 40.Roby KF, Taylor CC, Sweetwood JP, Cheng Y, Pace JL, et al. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis. 2000;21:585–591. doi: 10.1093/carcin/21.4.585. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Huang J, Yang N, Liang S, Barchetti A, et al. Integrative genomic analysis of protein kinase C (PKC) family identifies PKCiota as a biomarker and potential oncogene in ovarian carcinoma. Cancer Res. 2006;66:4627–4635. doi: 10.1158/0008-5472.CAN-05-4527. [DOI] [PubMed] [Google Scholar]
- 42.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 43.Zhao JJ, Gjoerup OV, Subramanian RR, Cheng Y, Chen W, et al. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3:483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 44.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao JJ, Cheng H, Jia S, Wang L, Gjoerup OV, et al. The p110alpha isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc Natl Acad Sci U S A. 2006;103:16296–16300. doi: 10.1073/pnas.0607899103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 47.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103:1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Yang N, Park JW, Katsaros D, Fracchioli S, et al. Tumor-derived vascular endothelial growth factor up-regulates angiopoietin-2 in host endothelium and destabilizes host vasculature, supporting angiogenesis in ovarian cancer. Cancer Res. 2003;63:3403–3412. [PubMed] [Google Scholar]
- 49.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(0.05 MB PDF)