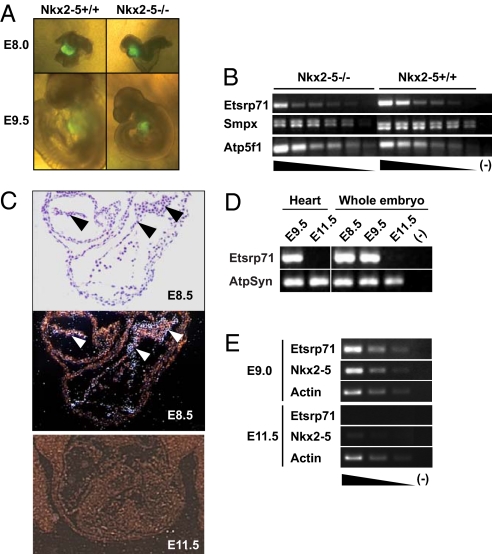
Fig. 1.
Transient expression of Etsrp71 in the endocardial/endothelial lineage is downregulated in Nkx2–5 null cardiac progenitors. (A) Six-kilobase Nkx2–5-EYFP Tg:Nkx2–5± and Nkx2–5± mice were mated to generate EYFP+ Nkx2–5+/+ and EYFP+ Nkx2–5−/− littermates at E8.0 and E9.5. Note that the WT and Nkx2–5 mutant embryos are indistinguishable at E8.0 while at E9.5 the Nkx2–5−/− embryo has severe growth retardation compared to the WT littermate. (B) Semiquantitative RT-PCR in 6-kb Nxk2–5-EYFP Tg:Nkx2–5 null vs. 6-kb Nxk2–5-EYFP Tg:WT cardiac progenitors. Note decreased Etsrp71 transcript expression in the Nkx2.5−/− cardiac progenitors vs. the WT controls. (C) In situ hybridization techniques for Etsrp 71 expression at E8.5. The bright (Top) and dark (Bottom) fields of the transverse section of an E8.5 heart are shown and arrowheads indicate endocardium. Note an absence of Etsrp71 expression in the E11.5 embryo. (D) RT-PCR analysis of Etsrp71 expression in the developing heart and embryo. RNA was extracted at the indicated stages from the developing heart and embryo to analyze the expression of Etsrp71. ATP synthase (ATPSyn) was used as a loading control. (E) Coexpression of Etsrp71 and Nkx2–5 in endothelial progenitor cells of developing embryos. Tie2-GFP+ cells were isolated from developing embryos using FACS for semiquantitative RT-PCR (25 cycles). Actin was used as a loading control and was analyzed after 15 cycles.