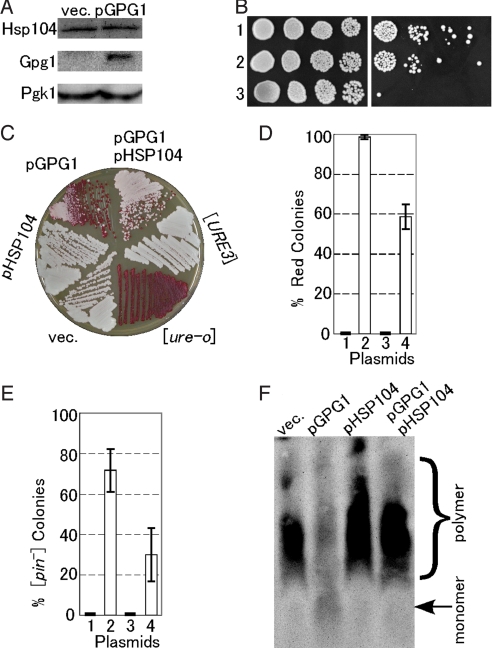
Fig. 3.
Effect of Gpg1 on expression and activity of Hsp104 and a compensatory effect of Hsp104 over-expression on prion elimination by Gpg1. (A) The cellular abundance of Hsp104. Equal amounts of total protein from whole cell lysates of NPK265 ([PSI+]) carrying plasmid pGPG1 or an empty vector (vec.) were separated by SDS/PAGE and immunoblotted with anti-Hsp104, -Gpg1, and -Pgk1 (control) antibodies. (B) Thermotolerance of Gpg1-overexpressing strain. Mid-log phase cultures of the indicated strains were incubated at 37 °C for 1 h to induce the heat-shock response and were then incubated for a further 20 min at 50 °C. The viability of heat-shocked cells (Right) was compared to untreated cells (Left) by 5-fold serial dilutions on YPD+ade agar. Cells were incubated at 30 °C for 2 days. Strains: 1, NPK265 carrying pGPG1; 2, NPK265 carrying an empty vector; 3, NPK339 (NPK265 derivative carrying hsp104Δ). (C) The DAL5-ADE2 reporter strain NPK302 ([URE3]) was transformed with pGPG1 and/or pHSP104, and the [URE3] state was monitored by colony color on YPD. (D) Semiquantitative estimation of the degree of [URE3] elimination. Strain NPK302 was transformed with the indicated plasmids. Fifty colonies from 3 independent transformations were grown on YPD plates; the frequency of red colonies was scored and the values are expressed with standard deviations. Plasmids: 1, empty vector; 2, pGPG1; 3, pHSP104; 4, pGPG1 plus pHSP104. (E) The [PIN+] strain (NPK200 [psi−]) transformants with pGPG1 and/or pHSP104 were transformed again with pRS414CUP1p-Rnq1-GFP, and the [PIN+] or [pin−] state was monitored by Rnq1-GFP fluorescent morphology as described in Fig. 2. Twenty transformant colonies, each from 3 independent transformations, were examined, scored for frequency of [pin−] colonies, and the values obtained are expressed with standard deviations. (F) The presence or absence of SDS-stable Rnq1 polymers in the above NPK200 transformants was analyzed by SDD-AGE as described in Fig. 2.