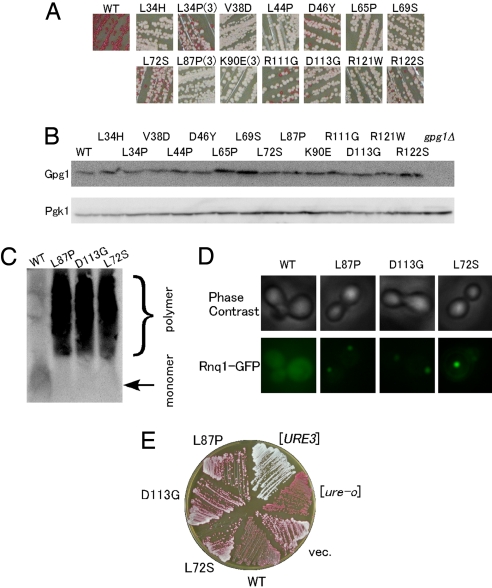
Fig. 5.
Loss-of-activity gpg1 mutants. (A) Color assay to determine the [PSI+]-eliminating ability of mutant Gpg1 proteins. NPK265 strain ([PSI+] ade1–14) was transformed with pGPG1 plasmids containing the indicated mutations, and transformants' color was monitored on YPD plates. Mutations isolated multiple times are marked by parentheses with the number of independent isolates, whereas the others are single isolates. WT denotes wild-type Gpg1. (B) The protein abundance of mutant Gpg1s. Lysates from the above transformants were examined by Western blotting as described in Fig. 1E. (C) SDD-AGE assay of [PIN+]-eliminating ability of mutant Gpg1 proteins. The formation of Rnq1 polymers in NPK200 ([PIN+]) transformants with pGPG1 plasmids carrying wild-type and the indicated gpg1 mutations was examined by SDD-AGE. (D) The same set of gpg1 mutant transformants as in (C) was transformed with pRS414CUP1p-Rnq1-GFP, and the [PIN+] state was examined by Rnq1-GFP fluorescence assay. (E) DAL5-ADE2-based color assay of [URE3]-eliminating activity of the gpg1 mutants. Experiments were performed as in Fig. 2D.