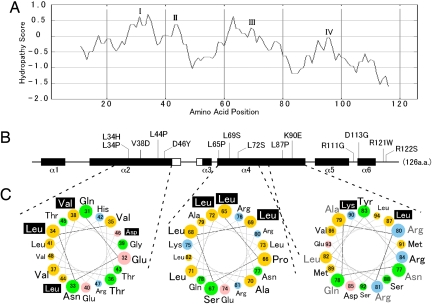
Fig. 6.
Protein architecture of Gpg1 and structural localization of loss-of-activity gpg1 mutations. (A) The hydropathy profile of Gpg1 generated by (http://www-personal.umich.edu/∼ino/blast.html). Representative hydrophobic regions are named I–IV. (B) Secondary protein structure prediction generated from the PSIPRED Protein Structure Prediction Server (http://bioinf.cs.ucl.ac.uk/psipred) (34). Open and closed boxes indicated β-sheets and α-helices (referred to α-1 through α-6), respectively, and bars indicate coils. Positions of amino acid substitutions are shown. Numbers represent the amino acid positions from the first Met codon. Note that the amino acid positions are shown in the same scale in (A) and (B) for comparison. (C) Helical wheel projection of residues in α-2 and α-4 of Gpg1. Residues 31–48 (helix α-2) and 65–94 (helix α-4) of Gpg1 are shown in a helical wheel projection created with the WheelApp applet (http://cti.itc.Virginia.EDU/∼cmg/Demo/wheel/wheelApp.html). Residues 65–94 (helix α-4) are split into 2 wheels in the same orientation: Note that residues 77–82 (gray, Right) are duplicated in both wheel projections in the same position. Acidic residues are shown in pink, basic residues in blue, neutral (uncharged polar) amino acids in green, and hydrophobic (nonpolar) amino acids in yellow. Loss-of-activity residues are designated with white text shadowed in black.