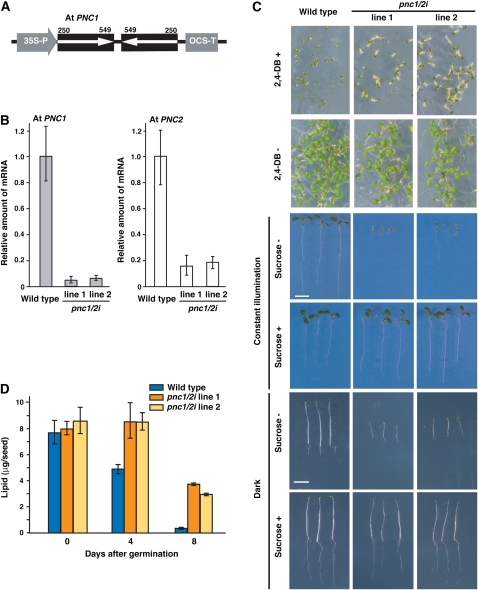
Figure 8.
Generation and Phenotypes of the pnc1/2i Knockdown Mutants.
(A) Transgene construction for generation of the pnc1/2i knockdown mutants in Arabidopsis. A partial cDNA fragment (250 to 549 bp) was derived from At PNC1. 35S-P is the cauliflower mosaic virus 35S promoter. OCS-T is octopin synthase terminator.
(B) The amounts of At PNC1 (gray bars) and At PNC2 (white bars) mRNA were determined by quantitative RT-PCR using seedlings grown for 3 d in the dark. The seedlings used were individual pnc1/2i mutants, line 1 and line 2, and the wild type. The data were normalized with respect to β-actin, and the amount of mRNA in wild-type seedlings on day 3 was normalized to 1.0. Error bars indicate se values of three independent experiments.
(C) Effects of 2,4-DB and sucrose on the growth of the knockdown mutant. Wild-type and pnc1/2i were grown for 7 d on growth medium with (+) or without (−) 0.35 μg/mL 2,4-DB under constant illumination. Wild-type and pnc1/2i were grown for 7 d on growth medium with (+) or without (−) sucrose under constant illumination or in the dark. Bars = 5 mm.
(D) The knockdown mutant exhibits defective degradation of seed lipid reserves during postgerminative growth. The amounts of reserved lipids in wild-type (blue bars) and pnc1/2i mutants (orange and yellow bars) were determined during postgerminative growth. Samples were prepared dry seeds (zero time) and etiolated seedlings. Error bars indicate se values of three or four independent experiments.