Abstract
Correlation of quantities of transcriptional activators and repressors with the mRNA output of target genes is a central issue for modeling gene regulation. In multicellular organisms, both spatial and temporal differences in gene expression must be taken into account; this can be achieved by use of in situ hybridization followed by confocal laser scanning microscopy (CLSM). Here we present a method to correlate the protein levels of the short-range repressor Giant with lacZ mRNA produced by reporter genes using images of Drosophila blastoderm embryos taken by CLSM. The image stacks from CLSM are processed using a semiautomatic algorithm to produce correlations between the repressor levels and lacZ mRNA reporter genes. We show that signals derived from CLSM are proportional to actual mRNA levels. Our analysis reveals that a suggested parabolic form of the background fluorescence in confocal images of early Drosophila embryos is evident most prominently in flattened specimens, with intact embryos exhibiting a more linear background. The data extraction described in this paper is primarily conceived for analysis of synthetic reporter genes that are designed to decipher cis-regulatory grammar, but the techniques are generalizable for quantitative analysis of other engineered or endogenous genes in embryos.
Introduction
Complex patterns of gene expression underlie the development of multicellular organisms, and capturing the spatial and temporal information of gene expression is critical for modeling gene regulatory networks. Microarrays or qPCR reactions can provide quantitative information, but these methods usually lack spatial information. On the other hand, fluorescent in situ hybridizations provide spatial information about gene expression and have the possibility of providing quantitative information as well. Recent studies have relied on such in situ techniques to quantitate the levels of nuclear proteins in the Drosophila embryo for the purposes of modeling.1,2
Because of its extensively researched genetic network, the Drosophila embryo provides one of the best-characterized systems for modeling transcriptional regulation. Transcription factors encoded by the maternal, gap, and pair rule genes form a regulatory network, whose interactions have been carefully described in molecular studies.3 A comprehensive mathematical description of this system still eludes us; however, in recent years several studies have modeled parts of this system.1,2,4,5 In most cases, confocal images of early Drosophila embryos were used to provide data on levels of transcription factors in the nucleus. One study reported a straightforward approach in which levels of regulatory proteins in the nucleus were related to protein levels of downstream targets, while other studies have focused on quantitative descriptions of mRNA levels in the embryo.6–8 A later study took a further step in correlating nuclear transcription factor levels to mRNA levels of a target gene but did not fully explore parameters and methods required for this analysis.4 Here, we report such a method that correlates the level of transcription factor to level of reporter gene mRNA, as a basis for mathematical modeling of gene regulatory elements.
Key to quantitative assessment of gene expression levels is information about the proportionality of signal read out to the actual levels of mRNA and protein. This relationship has been insufficiently tested; therefore, we examine this issue using gene dosage studies and independent mRNA measurements. Background (in this case, nonspecific fluorescence) is another central issue in many biological data analyses. A simple approach is to apply uniform background subtraction from the data. It has previously been noted that the background for fluorescently stained Drosophila embryos can be represented as a paraboloid function.9 Our study suggests that fluorescence background from undistorted embryos is not very paraboloidal, but as samples are flattened, a paraboloidal background becomes more evident.
This methodology appears to provide the correct basis for quantitative modeling approaches that utilize empirically established gene regulatory surfaces to facilitate parameter fitting. Such quantitative approaches will provide the tools to discover important regulatory information in genomic data sets, as well as lay a foundation for design of engineered transcriptional elements.
Materials and Methods
Immunofluorescent in situ hybridization
Embryos were collected and fixed as previously described.10 Immunofluorescent in situ hybridization was done essentially as previously described with some modifications.6,11 All washes were done in 1.0 mL. Fixed embryos stored at −20°C in methanol were briefly washed six times with 100% ethanol and then with xylene for 1 h. About 50 μL of embryos was transferred into individual microfuge tubes and washed four times with methanol-phosphate buffer 0.1%-Tween80 ([PBT; 1.37 M NaCl, 43 mM Na2HPO4, and 14 mM NaH2PO4], 1:1, v/v ratio) and then with PBT four times, each for 3 min with continuous rocking. Embryos were washed in PBT with hybridization solution (50% formamide, 5× SSC [3 M NaCl and 0.3 M Na-citrate], 100 μg/mL sonicated salmon sperm DNA, 50 μg/mL heparin, and 0.1% Tween80 [1:1, v/v ratio]) for 10 min, and then briefly in 100% hybridization solution for 2 min. New hybridization solution was added, and the tubes were placed for 1 h in a water bath at 55°C. Antisense RNA probes of digU-labeled lacZ were heated in 50 μL hybridization solution at 80°C for 3 min and directly placed on ice for 1 min; then prehybridization solution was completely removed, the probe in 50 μL of hybridization solution was added to each tube, and tubes were incubated at 55°C for 18–20 h. After incubation, 1 mL of 55°C hybridization solution was added to each tube; all tubes were rocked at room temperature for 1 min, hybridization solution was changed, and tubes were incubated for another 1 h at 55°C, followed by four washes with hybridization solution for 15 min each at 55°C and with hybridization solution and PBT (1:1, v/v ratio) two times at room temperature for 15 min. Five more washes were done with PBT for 10 min with rocking at room temperature. The embryos were washed with a blocking solution 1.0% casein in maleic acid buffer (Western Blocking Reagent; Roche, Indianapolis, IN) with PBT (1:1, v/v ratio). About 0.5 mL PBT and blocking solution (1:1, v/v) containing primary antibodies (2.2 μL of 1:250 dilution of mouse anti-DigU; Roche) and 0.75 μL of 1:800 dilution of rabbit anti-Giant (antibodies are a gift from Reinitz Lab12) was added, and the tubes were rocked at 4°C overnight. Tubes were washed four times each with PBT for 15 min at room temperature. About 0.4 mL of PBT and 0.5% casein blocking reagent (1:1) containing 8.0 μL of secondary antibodies—goat anti-mouse conjugated to Alexa 555 for detection of lacZ mRNA and chicken anti-rabbit conjugated to Alexa 488 for detection of the Giant protein (Molecular Probes, Eugene, OR) preabsorbed against fixed yw embryos—were added to each vial, and the tubes were covered with aluminum foil to protect them from light and incubated overnight at 4°C. Embryos were then washed with PBT four times at room temperature for 5 min with rocking, and washed in glycerol + PBT (7:3, v/v ratio) for 2 h until the embryos settled to the bottom of the tubes. The embryos were then resuspended in 0.4 mL glycerol + PBT (9:1 ratio) and 0.2 mL of Permafluor Mountant Medium 434990 (Thermo Electron Corporation, Pittsburgh, PA), mounted labeled slides, and covered with large rectangular Corning cover slips (No. 1.5; 24 × 50 mm). The slides were protected from light and stored flat at room temperature until they were imaged.
Sequences for reporter genes
The reporter constructs analyzed here are synthetic; Twist and Dorsal activator binding sites were previously characterized in lacZ reporter genes in the embryo.13 High-affinity Giant binding sites derived from the Kruppel promoter were previously characterized.14 The 25 bp neutral spacer used lacks high-affinity sites to blastoderm transcriptional regulatory proteins. Two reporters are tested here, both containing a core of two Twist sites 5′ of two Dorsal sites. A pair of Giant motifs is located either 31 or 131 bp 5′ of the activators. Binding sites are capitalized, Giant sites are italicized, Twist sites are bold, and Dorsal sites are underlined.
2gt.25.2T2D:
5′–gaattcTATGACGCAAGAatgcgactcgTATGACGCAAGAggatctggttagtaagctgtaaactgga tccCATATGttgagCATATGtctagaGGGATTTTCCCAaatcgaGGGAAAACCCAAccgcgg–3′
2gt.125.2T2D
5′–gaattcTATGACGCAAGAatgcgactcgTATGACGCAAGAggatctggttagtaagctgtaaactg gatctggttagtaagctgtaaactggatctggttagtaagctgtaaactggatctggttagtaagctgtaaactggatctggt tagtaagctgtaaactggatccCATATGttgagCATATGtctagaGGGATTTTCCCAaatcgaGGGAAAACCCAAccgcgg–3′.
Confocal laser scanning microscopy
An inverted confocal laser scanning microscopy (CLSM) Olympus Fluoview FV1000 (Olympus, Center Valley, PA) was used for capturing the confocal fluorescent images. For each scan of mounted embryos, the same microscope settings were employed to all images to simplify comparison of results. The argon laser (488 nm) was set at 5.0% and helium–neon laser (543 nm) was set at 25%. Emitted fluorescence from Alexa 488 and 555 was detected through a dichroic 405/488/543 and a BP505–525 filter for the green channel and a BP560–620 filter for the red channel. The pinhole was set to 105 μm (1.0 Airy unit), and the PMT detector, gain, and offset were 680, 1.0%, and 6% for both green and red channels. The PMT detector was adjusted in cases where the embryos showed saturation of signal intensities. Embryos were imaged at a scan speed of 6.51 s/scan, line filter equal 2 line Kalman filter, and 1.73-μm-thick Z-sections for a total of 21–30 slices. CLSM image data were stored as two separate stacks and projections of images for each channel. The section dimensions were 333 μm in length and width, and 1.73 μm in depth. Fluorescence pixels were recorded as 12-bit images and stored as TIFF files.
Determining proportionality of fluorescence intensity
To test whether immunofluorescent signals were proportional to the actual levels of protein or mRNA, we varied gene dosage of lacZ reporter or the Giant repressor, assuming that heterozygotes express at half the level of homozygotes. Male transgenic flies homozygous for a lacZ reporter gene regulated by the Giant repressor and Twist and Dorsal activators were crossed with virgin yellow white females to produce heterozygous embryos carrying a single copy of the lacZ gene. Homozygous lacZ reporter embryos were also collected. Two to 4 h old homozygous and heterozygous embryos were dechorionated, fixed, and analyzed for lacZ expression using immunofluorescent in situ hybridization. For RTqPCR mRNA analysis, dechorionated embryos were frozen in liquid nitrogen and stored at −80°C until needed. For extraction of total RNA, frozen embryos were dipped briefly in liquid nitrogen and macerated with a pestle in 500 μL of TriaZol reagent from Invitrogen (Carlsbad, CA) in a 1.5 mL microfuge tube. The extraction and subsequent DNase (Roche) treatment were performed according to the manufacturer's directions. To test for the presence of any DNA contamination, standard PCR was performed using primers specific for lacZ gene. RNA was spectrophotometrically quantified, and equivalent amounts of RNA from each sample were used for the reverse transcriptase reaction using the Superscript III enzyme. The RTqPCR-specific primer set for lacZ (F: 5′-CTGGGATCTGCCATTGTCAGA; R: 5′-TGGTGTGGGCCATAATTCAATT) was designed using Primer Express Software (ABI 7500 Prism). RTqPCR normalization and analysis were done as described previously.15,16 Primer sets of actin (F: 5′-CGCGGTTACTCTTTCACCA; R: 5′-GCCATCTCCTGCTCAAAGTC) and 28S rRNA (F: 5′-GATGCCGCGCTAGTTACAT; R: 5′-GCTGCTCAACCACTTACAACAC) were used for normalization.
For analysis of Giant protein levels, a gtX11 stock was obtained from the Bloomington Stock Center.17 gtX11 virgins were out crossed with wild-type male flies to obtain male hemizygous gtX11 mutant embryos, gtX11 and heterozygous female embryos, and wild-type female embryos.
Image processing prior to lacZ mRNA and Giant repressor measurement
In this study, Image J software (http://rsbwebnih.gov/ij/) and MATLAB software (2007a, The Mathworks, Natick, MA) were used. Image J software was used to extract the fluorescent intensity levels of lacZ and Giant from each confocal fluorescent image. Software implementing the algorithms described below was written in MATLAB and is available upon request. Approximately 18–21 nonoverlapping 1.73-μm-deep optical sections were taken from each embryo, providing a 1024 × 1024 pixel 12-bit image representing the top half of the embryo. The slices were projected by taking maximum intensity for each pixel along the z-axis, and the images were converted to 8-bit images. For flatter embryos analyzed in a previous study, the average of two adjacent sections was used, but with our more three-dimensional imaging, this approach is not valid because for most sections a portion of the central part of the embryo containing only yolk is included.6
To position images in a uniform horizontal manner, masks were used to rotate the images as described in Janssens et al.6 A mask is created for the projected images by using the threshold value that is taken from outside of the embryo. The convex hull of the embryo boundary is taken, the inside of the convex hull is set to 255, and the outside is set to 0. The rotated images were checked to compare the alignments of the two rotated channels. Images were flipped upside-down or left–right to align uniformly on the anterior–posterior and dorsal–ventral axes. To remove extraneous portions of the image outside the embryo, the rotated images were cropped to a minimal canvas size. The images were not changed to a uniform size in order to avoid interpolation of the data, which might have varying effects for different embryos. Subsequent to these steps, perimeter pixels of the embryo were found using the mask.
In the next step, embryo boundaries, lacZ mRNA data, and Giant repressor protein data were used to decide where to set boxes that are approximately the size of an average cell. The embryos are at this point incompletely cellularized, but separate nuclei are clearly discernible. Positioning of the boxes is explained in “Methods for data collection” section. The Giant protein and lacZ mRNA levels were averaged inside those boxes and plotted in two-dimensional (2D) space.
Comparison of imaging to data from other databases
A variety of methods have been applied to obtain spatially resolved mRNA and protein information from Drosophila embryos. Our CLSM images of Giant were obtained using antibody staining and one-photon imaging, but embryos were not flattened as in a previous study.1 Alternatively, mRNA quantitation from nondeformed embryos has been acquired by two-photon imaging.8 We compared qualitative features of our Giant images with these data sets to see if general trends were consistent. As an example, we observed in some of the images lower apparent levels of Giant protein in the ventral and dorsal regions of the embryo. Giant images were accessed from two other databases: the Berkeley Drosophila Transcription Network Project (BDTNP) and the Database of Segmentation Gene Expression in Drosophila (FlyEx). mRNA levels in actual embryos from the BDTNP and protein levels from an average virtual embryo image from the FlyEx were used.8,18 Overall relative differences in intensities between anterior and posterior levels of Giant protein and mRNA were observed in all three data sets, as well as the relative lower levels of Giant in ventral regions of the anterior stripe (data not shown). Despite differences in imaging, these methods appear to capture the same essential features of this gene's expression.
Results
Background
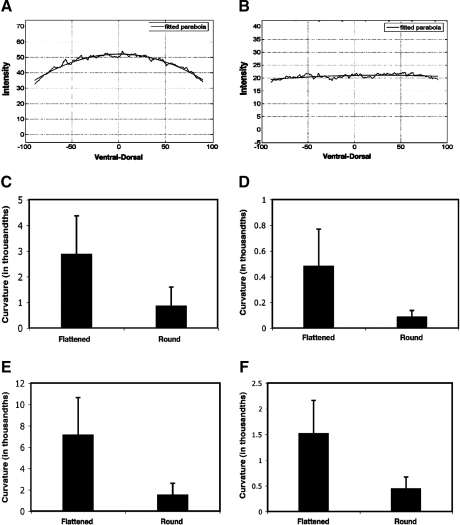
A critical issue in quantitative measurement of gene expression is background subtraction. In a previous study, it was noted that the background coming from nonspecific binding of a variety of primary and secondary antibodies to Drosophila embryos can be approximated by a paraboloid.9 In that study, embryos were flattened (by the weight of a coverslip and using reduced amounts of mounting solution) so that a significant fraction of nuclei of the embryo could be captured in two 2 μm sections. In our system, we do not distort the embryo; thus, we tested whether this parabolic background relationship still applied. By assessing fluorescence in two different channels in embryos lacking a lacZ reporter gene, as well as in portions of embryos devoid of Giant protein (45–55% egg length), we found that the background often did not show a paraboloid shape (Fig. 1A, B). One evident difference between our methods and those described in Myasnikova et al.9 is the degree of flatness of the embryos. We imaged embryos of successively flatter proportions and analyzed background levels as a function of flatness (Fig. 1C–F). Embryo flatness was judged by the number of 1.73 μm slices required to reach the center of the embryo. Embryos that were flattened by a weighted coverslip had radial thicknesses ranging from 14 to 23 μm, while rounder embryos had thicknesses of 24–33 μm. This level of flattening is not as pronounced as that described in Myasnikova et al.,9 where almost the entire upper half of the embryo can be scanned in two 2 μm slices, but a clear trend emerged. The background has a distinct tendency to be more paraboloidal with flatter embryos, perhaps because unique light scattering properties of flat embryo sections contribute to a nonlinear background. Correlation between flatness of the embryos and curvature of the background intensity is measured by Pearson's correlation coefficients. For lacZ images, the Pearson's correlation coefficients between flatness of the embryo and curvature of the background curve are −0.5 (p = 0.006) (ventral–dorsal) and −0.6 (p <0.001) (anterior–posterior). When this relationship is measured as the correlation between embryo flatness and the natural log of the curvature, the correlation coefficients are −0.6 (p < 0.001) (ventral–dorsal) and −0.7 (p < 0.001) (anterior–posterior). The relationship between flatness of the embryo and curvature of the background curve can be explained better with nonlinear functions. The main point here is that parabolic background cannot be assumed without taking the geometry of embryo into account. We also noted that for some embryos, inhomogeneously distributed background was evident in both channels (488 and 555 nm), suggesting that specific structural features of embryos can affect background (data not shown).
FIG. 1.
Parabolic and nonparabolic background forms. Representative parabolic (A) and flat (B) backgrounds. Data were taken from the middle 45–55% egg length of embryos. In general, we observed parabolic background for flatter embryos. Embryo cross-sectional radii were measured and classified as flattened (14–23 μm) or round (24–33 μm). Considerable variability in curvature was noted, but flatter embryos tended to exhibit higher curvature overall. Data were obtained for Giant imaging ventral–dorsal (C) and anterior–posterior (D), and lacZ imaging ventral–dorsal (E) and anterior–posterior (F). Parabolicity of background is measured by curvature of the parabola fit to curve, with apex generally at center of embryo cross section. For anterior–posterior Giant background measurements, young gtX11 mutants expressing virtually no detectable Giant protein were used. For lacZ background imaging embryos without the reporter gene were utilized.
Fluorescent quantitation of mRNA and protein
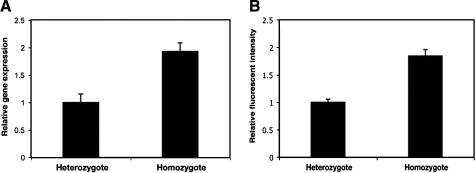
Quantitative measurements of transcription factor levels and mRNA inputs are essential for modeling gene regulation, yet surprisingly few studies relying on CLSM have independently tested whether the signals thus obtained exhibit strongly nonlinear effects. A quantitative study of Drosophila mRNA levels made a single correlation between knirps mRNA and Knirps protein obtained by CLSM as a test for proportionality, while other studies have quantitated levels of green fluorescent protein as a proxy for transcript levels per cell.8,19 We concluded that more rigorous independent measurements are essential for testing the validity of our quantitation. In the first case of mRNA detection, we varied the gene dose of a lacZ reporter expressed in ventral regions of the embryo and compared levels of mRNA by RTqPCR and CLSM. The RTqPCR results showed that the relative amount of lacZ mRNA was 1.0 in heterozygotes compared to 1.92 in homozygotes (Fig. 2A). In comparison, fluorescent intensities for lacZ heterozygous compared to homozygous embryos were 1 to 1.85 (Fig. 2B). This result suggests that both methods are responding to estimated twofold differences in mRNA in a similar manner, and that the fluorescent intensities do provide a reasonable proxy for actual mRNA levels.
FIG. 2.
Proportionality of signal to mRNA levels. The average relative amounts of lacZ mRNA in heterozygous and homozygous transgenic embryos were measured by RTqPCR analysis (A) and by immunofluorescent in situ hybridization and CLSM (B). Quantitation of mRNA levels by RTqPCR is representative of two biological assays; error bars indicate standard deviation from five technical replicates. Fluorescent imaging of 39 embryos were quantitated in (B).
Regarding protein measurement, previous studies have analyzed relative expression levels of a number of regulatory proteins in the Drosophila embryo by CLSM, but to our knowledge the proportionality of these measurements to actual protein has not been assessed.1 We tested the measurement of protein by comparing the values of Giant expression in embryos derived from an outcross of gtXII heterozygotes to a wild-type strain (Fig. 3). gt is located on chromosome 1; thus, male hemizygotes carrying the mutant allele would be expected to express the lowest levels of the protein, and female heterozygotes would have intermediate levels. The allele used, gtX11, has been reported to express low levels of protein, and consistent with this observation, no mid-blastoderm embryos entirely lacked Giant expression.
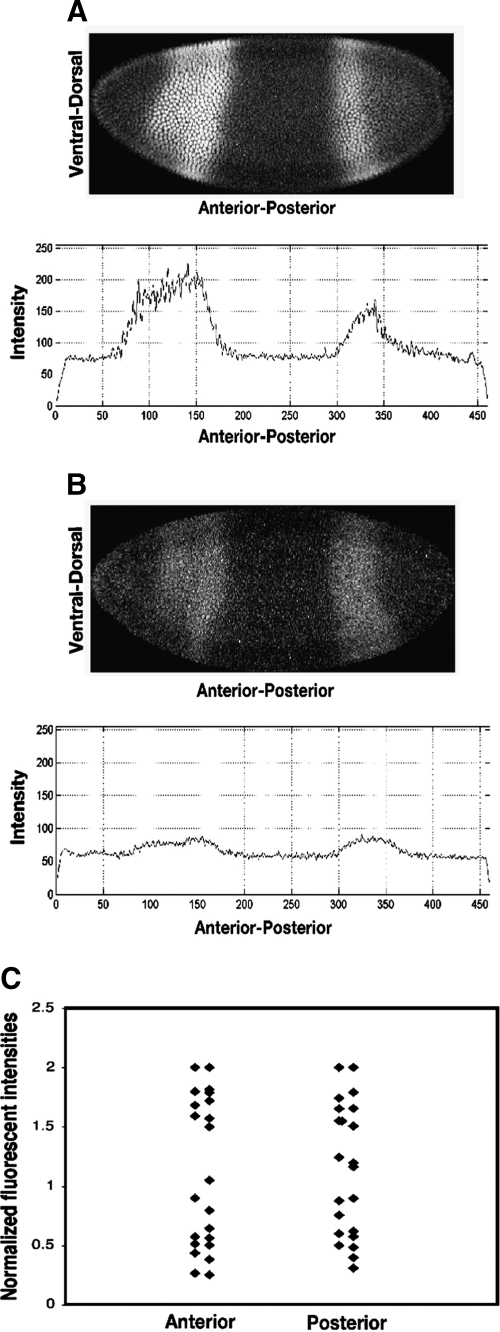
FIG. 3.
Proportionality of signal to Giant protein levels. Embryos with apparently normal intensity of Giant (A) and low intensity of Giant (B) were observed for age-matched embryos. Normalized levels of 22 background-subtracted Giant images were plotted, and a roughly 10-fold range of signal intensities were observed (C). Overlapping data points were separated into two columns for clarity for the anterior and posterior stripe.
Embryos were stage matched to allow direct comparison of protein levels. Embryos with apparently normal intensity of Giant signal were observed alongside of age-matched embryos with very low Giant levels (Fig. 3A, B); those differences are not likely to be merely a function of overall staining efficiency because background levels were fairly constant and such large differences are not usually seen when imaging wild-type embryos. When background subtracted, normalized levels of Giant were plotted for a set of 22 images, a roughly 10-fold range of signal intensities were observed (Fig. 3C). The values did not fall into three obviously discrete clusters, corresponding to hemizygous null, heterozygous, and wild-type backgrounds, but were rather continuously distributed with some degree of over representation at higher and lower values. This effect may be due to nonlinearity of the fluorescent readout, which would produce some compression at one end of the spectrum, variability in detection or even expression of Giant from embryo to embryo, or some combination of these effects. We can conclude that this method does permit an apparent dynamic detection range of at least 10-fold sufficient to capture the total dynamic variation reported for Giant in the blastoderm embryo, but clearly a more detailed comparison of signal proportionality is warranted.1 In light of our mRNA results, it appears that fluorescent detection can be an appropriate proxy for in situ levels of biomolecules, but the proportionality of read out must be established for each set of reagents.
Methods for data collection
To understand the functional relationship between transcriptional activator and repressor levels and mRNA output, we need to create gene regulatory maps that describe the quantitative relationship between these elements.20 We created a series of lacZ reporter genes regulated by the Giant transcriptional repressor and the Dorsal and Twist activators. These reporter genes are active in ventral regions of the embryo, and expression is interrupted in areas where Giant is expressed, depending on the arrangement of the binding sites (Fig. 4A, B). We directly measure levels of the Giant repressor protein as an input value. The spatially and temporally varying levels of this protein provide in each embryo an entire set of values relevant to a gene regulatory map. Activator levels can be similarly measured, and in the case of Dorsal and Twist, these activators vary along the dorsal to ventral axis.2 We focus on gene modules that test varying features of repressor binding sites, while holding Dorsal and Twist sites constant. Dynamic and spatially heterogeneous protein levels of transcriptional regulators make it imperative that we compare lacZ levels with corresponding levels of regulatory factors in nearby nuclei from which the mRNA originates.
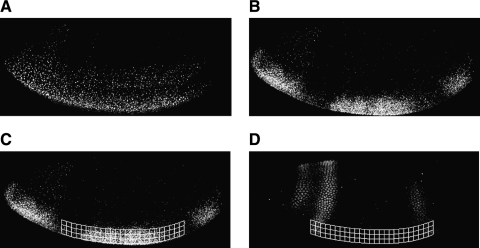
FIG. 4.
Sampling of integrated mRNA and Giant data. lacZ reporter constructs were introduced into Drosophila by germline transformation and mRNA detected by fluorescent imaging. Dorsal and Twist activators drive lacZ gene expression in ventral regions (A). When regulated by Giant, gaps in this pattern are evident (B). A sampling mesh is imposed upon the background-subtracted image, and values for Giant and lacZ are collected (C, D). The mesh size here is exaggerated for clarity. Sampling proceeds from anterior region of anterior stripe to posterior region of posterior stripe.
Background
Background intensity for the Giant channel is calculated by averaging the data from the middle (50–60% egg length) of the embryo along the ventral–dorsal axis. In cases where the curve shows linear behavior, the average of this middle stripe can be subtracted from the whole embryo, and if it shows parabolic behavior, then the background can be calculated and subtracted as previously described.9 Background intensity for the lacZ channel can be calculated similarly by using the dorsal parts of the embryo, where no lacZ is present.
Binning
A pixel by pixel comparison between Giant, found in the nucleus, and the lacZ mRNA channel is not applicable because the mRNA accumulates in the spatially separated cytoplasm. In addition, we observe that lacZ mRNA accumulates in a punctuate pattern (a function of the specific reporter, as other mRNAs show a smoother, nonnuclear pattern; data not shown). This problem can be solved by measuring the approximate size of a cell in confocal images and covering the region of interest by boxes of the size of an average cell size. A 10 × 10 pixel box is the average cell size for our images.
Relevant areas of data collection
We then collect a series of data points representing the lacZ gene output in regions with similar levels of activators but varying levels of Giant protein. When not regulated by Giant, the lacZ expression pattern is almost constant from approximately 20–80% egg length (anterior–posterior) in the ventral part of the embryo (Fig. 4A). When Giant is effective at repressing the lacZ reporter, gaps in this pattern are seen in anterior and posterior regions (Fig. 4B). It has been reported that Twist mRNA levels are not uniform from anterior to posterior,21 but this variation does not seem to be reflected in the output of our reporters, which are activated by Twist and Dorsal in a fairly uniform pattern in the mid-blastoderm stage. For this reason, in the regions that are exactly parallel to the ventral boundary, we assume that the combined activator effect is not changing for our synthetic enhancer constructs. We therefore measure correlated lacZ and Giant levels in slices that track along the ventral aspect of the embryo (Fig. 4C, D). Curved slices, 10 pixels in width, are taken from ventral part of the embryo along anterior to posterior, following the boundary of the embryo. Each slice is divided into 10 × 10 pixel boxes along anterior to posterior to derive mean values for lacZ and Giant.
To accommodate images of embryos that are rotated around the anterior–posterior axis to different degrees, exposing more or less of the ventral surface where Dorsal/Twist are active, we vary the number of slices applied to each embryo so that our lacZ mRNA and Giant measurements reflect areas with nearly constant activator levels. To determine the number of slices used, we measure lacZ levels in central regions (50–60% egg length) that are not subject to Giant regulation and extend slices from ventral to dorsal until activator levels are determined to be limiting. This is accomplished by measuring lacZ expression in the 50–60% egg length portion of the embryo and averaging along the anterior–posterior axis, producing a plot that describes how lacZ is changing from ventral to dorsal. This data has a peak value at the ventral side of the embryo and decreases from ventral to dorsal. The slices start from the peak level of lacZ expression and end at 50% of the maximal lacZ expression.
Because the ventral lacZ expression is also affected by vector-mediated activation in anterior regions, as well as Torso-mediated regulation of Dorsal22 in the poles, our slices start from a position of half of the maximal Giant intensity on the anterior side of the major anterior stripe, to half of the maximal Giant intensity of the posterior side of the posterior Giant stripe. The selected region still includes most of the region of Giant expression and removes portions of the data subject to extraneous influences (Fig. 4C, D).
Normalization and regulatory plots
The quantitation described above focuses on the lacZ levels as a function of the Giant concentration (Fig. 5). The Giant data is normalized by dividing the 2–4 h old embryo time interval studied here into eight segments as described in Jaeger et al.1 The average levels of the protein in each interval are used to normalize the Giant signals. In this way, comparisons of lacZ expression to relative Giant levels in a given embryo can be combined with data from other embryos into larger data sets. Because Dorsal and Twist activator proteins are relatively constant, each embryo can be normalized by dividing by its maximum intensity value. These normalized expression levels of Giant protein and lacZ mRNA are used to plot expression surfaces, showing the repressor signal and activator signal versus output (Fig. 5). For the visualization shown here, the data is presented as 2D plots, in which each box represents one slice from the ventral region of the embryo. As expected, with increasing Giant levels or decreasing activator levels, the lacZ signal is reduced. The resulting plot represents 2D slices of a gene regulatory surface that is unique to the specific regulatory region of interest.
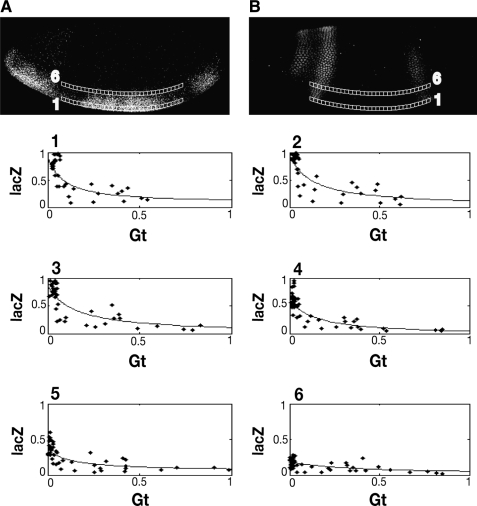
FIG. 5.
Gene regulatory representations from Giant-regulated lacZ reporter gene. To generate gene regulatory plots, data derived from slices (lacZ mRNA channel [A] and Giant protein channel [B]) are plotted in a 2D space. Only two slices are shown for clarity. The plots numbered 1–6 show how robust lacZ levels present in ventral portions of the embryo are repressed in regions containing peak Giant levels such as in plot 1–4. In more dorsally located regions (plot 5–6), limiting levels of activators reduce lacZ expression regardless of Giant levels. The overlaid lines, obtained by fitting the data with a rational function, show the trend of the relation between levels of Giant and lacZ.
Previous studies have demonstrated that the Giant protein can repress transgenes regulated by Dorsal and Twist, but the characterization of this relationship has never gone beyond a qualitative nature.13 To characterize the quality of the data obtained from this system and assess its utility for quantitative modeling, we measured the correlation between [Gt] and ln ([lacZ]) using the Pearson's correlation coefficient. For the 2gt.25.2T2D construct showing regulation by Giant, correlation coefficients for slice 1 to slice 6 are −0.74, −0.78, −0.82, −0.85, −0.65, and −0.67. The p-values for these correlation levels are all less than 0.0001, indicating statistical significance.
Discussion
We describe a method to correlate the transcription factors and mRNA output for gene modeling purposes. We show that the levels of lacZ mRNA and potentially the transcriptional repressor protein Giant are proportional to fluorescent intensities, a critical basis for quantitative modeling. Our analysis also reveals that a suggested parabolic form of the background fluorescence in confocal images of early Drosophila embryos is evident most prominently in flattened specimens, with intact embryos exhibiting a more linear background. After appropriate background subtraction and normalization, these data are amenable to representation of gene regulatory surfaces that permit creation and validation of quantitative models of gene expression. In this way, we have constructed the foundations for modeling a cis-regulatory grammar that applies to an important set of transcriptional regulators in the Drosophila blastoderm embryo. More generally, the image and data analysis techniques described in this paper can be generalized to understand the quantitative function of endogenous genes or gene networks in the Drosophila embryo or other well-characterized systems. Such an approach may also prove useful for engineered transcriptional regulatory elements employed for targeted expression of genes in therapeutic settings.
Acknowledgments
We thank Melinda Frame for assistance with confocal imaging, and members of the Arnosti lab for discussions and comments on the manuscript. This study was supported by the MSU Foundation, the MSU Quantitative Biology and Modeling Initiative, and NIH GM56976 to David N. Arnosti.
References
- 1.Jaeger J.Surkova S.Blagov M.Janssens H.Kosman D.Kozlov K.N. Manu, Myasnikova E.Vanario-Alonso C.E.Samsonova M.Sharp D.H.Reinitz J.Dynamic control of positional information in the early Drosophila blastoderm Nature 430368.2004 [DOI] [PubMed] [Google Scholar]
- 2.Zinzen R.P. Senger K. Levine M. Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol. 2006;16:1358. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 3.Rivera-Pomar R. Jackle H. From gradients to stripes in Drosophila embryogenesis: filling in the gaps. Trends Genet. 1996;12:478. doi: 10.1016/0168-9525(96)10044-5. [DOI] [PubMed] [Google Scholar]
- 4.Janssens H. Hou S. Jaeger J. Kim A.-R. Myasnikova E. Sharp D. Reinitz J. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat Genet. 2006;38:1159. doi: 10.1038/ng1886. [DOI] [PubMed] [Google Scholar]
- 5.Segal E. Raveh-Sadka T. Schroeder M. Unnerstall U. Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 6.Janssens H. Kosman D. Vanario-Alonso C.E. Jaeger J. Samsonova M. Reinitz J. A high-throughput method for quantifying gene expression data from early Drosophila embryos. Dev Genes Evol. 2005;215:374. doi: 10.1007/s00427-005-0484-y. [DOI] [PubMed] [Google Scholar]
- 7.Tomancak P. Beaton A. Weiszmann R. Kwan E. Shu S. Lewis S.E. Richards S. Ashburner M. Hartenstein V. Celniker S.E. Rubin G.M. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3:88. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luengo Hendriks C.L. Keranen S.V.E. Fowlkes C.C. Simirenko L. Weber G.H. DePace A.H. Henriquez C. Kaszuba D.W. Hamann B. Eisen M.B. Malik J. Sudar D. Biggin M.D. Knowles D.W. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution 1: data acquisition pipeline. Genome Biol. 2006;7:R123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myasnikova E. Samsonova M. Kosman D. Reinitz J. Removal of background signal from in situ data on the expression of segmentation genes in Drosophila. Dev Genes Evol. 2005;215:320. doi: 10.1007/s00427-005-0472-2. [DOI] [PubMed] [Google Scholar]
- 10.Small S. Blair A. Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:3147. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosman D. Mizutani C.M. Lemons D. Cox W.G. McGinnis W. Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- 12.Kosman K. Small S. Reinitz J. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev Genes Evol. 1998;208:290. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- 13.Szymanski P. Levine M. Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. EMBO J. 1995;14:2229. doi: 10.1002/j.1460-2075.1995.tb07217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hewitt G.F. Strunk B.S. Margulies C. Priputin T. Wang X.D. Amey R. Pabst B.A. Kosman D. Reinitz J. Arnosti D.N. Transcriptional repression by the Drosophila giant protein: cis element positioning provides an alternative means of interpreting an effector gradient. Development. 1999;126:1201. doi: 10.1242/dev.126.6.1201. [DOI] [PubMed] [Google Scholar]
- 15.Huggett J. Dhehada K. Bustin S. Zumla A. Real-time RT-PCR normalization; strategies and considerations. Genes Immun. 2005;6:279. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 16.Vandesompele J. de Preter K. Pattyn F. Poppe B. van Roy N. de Paepe A. Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:34. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eldon E. Pirrotta V. Interactions of the Drosophila gap gene giant with maternal and zygotic pattern-forming genes. Development. 1991;111:367. doi: 10.1242/dev.111.2.367. [DOI] [PubMed] [Google Scholar]
- 18.Myasnikova E. Samsonova A. Kozlov K. Samsonova M. Reinitz J. Registration of the expression patterns of Drosophila segmentation genes by two independent methods. Bioinformatics. 2001;17:3. doi: 10.1093/bioinformatics/17.1.3. [DOI] [PubMed] [Google Scholar]
- 19.Damle S. Hanser B. Davidson E.H. Fraser S.E. Confocal quantification of cis-regulatory reporter gene expression in living sea urchin. Dev Biol. 2006;299:543. doi: 10.1016/j.ydbio.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 20.Setty Y. Mayo A.E. Surette M.G. Alon U. Detailed map of cis-regulatory input function. PNAS. 2003;100:7702. doi: 10.1073/pnas.1230759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X. MacArthur S. Bourgon R. Nix D. Pollard D.A. Iyer V.N. Hechmer A. Simirenko L. Stapleton M. Luengo Hendriks C.L. Chu H.C. Ogawa N. Inwood W. Sementchenko V. Beaton A. Weiszmann R. Celniker S.E. Knowles D.W. Gingeras T. Speed T.P. Eisen M.B. Biggin M.D. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rusch J. Levine M. Regulation of the dorsal morphogen by the Toll and torso signaling pathways: a receptor tyrosine kinase selectively masks transcriptional repression. Genes Dev. 1994;8:1247. doi: 10.1101/gad.8.11.1247. [DOI] [PubMed] [Google Scholar]