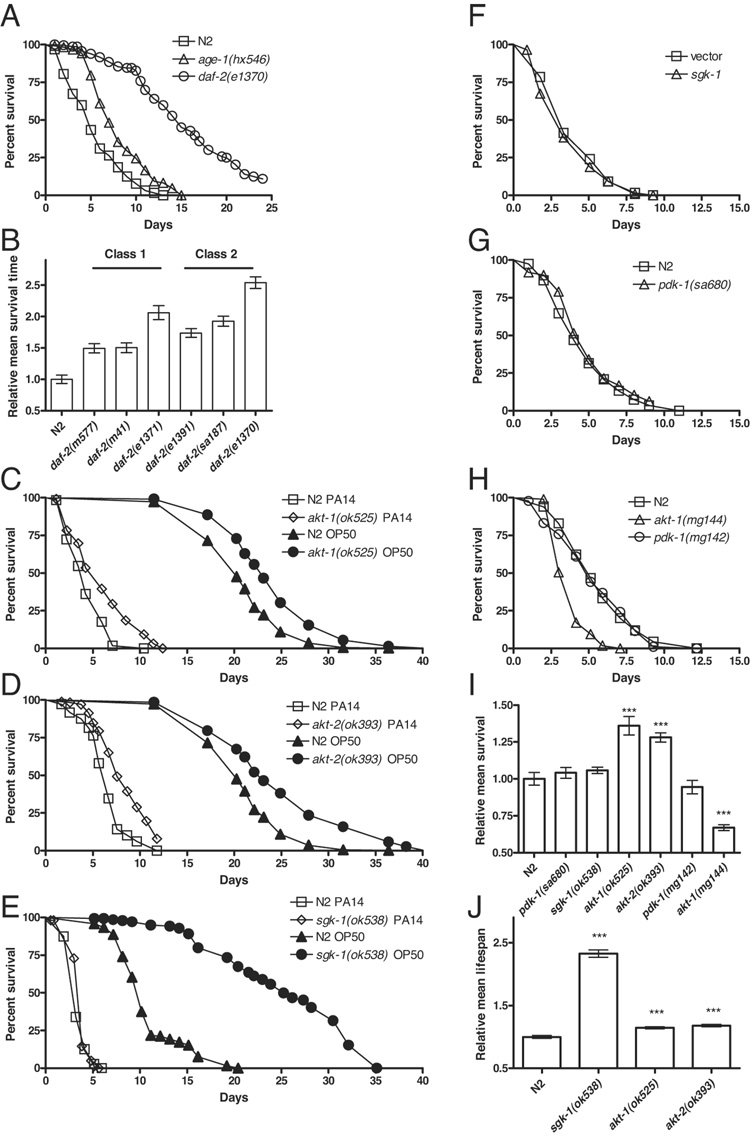
Figure 2. Aging and pathogen resistance are distinctly regulated by components of the insulin-like signaling pathway.
(A) The enhanced pathogen resistance of long-lived daf-2 and age-1 mutants is observed in sterile animals. Survival of sterile daf-2(e1370), age-1(hx546), and N2 adults on PA14 over time is shown. The survival of each strain was significantly different in pairwise comparisons (logrank, p < 0.0001). (B) Hypomorphic daf-2 mutants were resistant to PA14. daf-2(m577), daf-2(m41), daf-2(1371), daf-2(e1391), daf-2(sa187), and daf-2(e1370) were more resistant to pathogen than N2 (logrank, p < 0.0001). Of the daf-2 mutants tested, daf-2(e1370) was reproducibly the most resistant (logrank, p < 0.0001 in pairwise comparisons). Note that pathogen resistance varied more within than between phenotypic classes among daf-2 alleles (Gems et al. 1998). (C) akt-1(ok525) mutants were resistant to PA14 (logrank test, p < 0.0001; open symbols). They were also slightly, but significantly longer lived, with a mean lifespan of 115% compared to N2 (logrank p < 0.0001; closed symbols) when fed with UV-killed OP50 as 1-day old adults. (D) akt-2(ok393) mutants were resistant to PA14 (logrank test, p < 0.0001; open symbols). They were only slightly but significantly longer lived, with a mean lifespan of 118% compared to N2 (logrank p < 0.0001; closed symbols) when fed with UV-killed OP50 as 1-day old adults. (E) sgk-1(ok583) mutants were not significantly different than N2 in resistance to PA14 (logrank test, p = 0.11). sgk-1(ok538) mutants exhibited a mean lifespan of 230% compared to N2 (logrank, p < 0.0001). Lifespan of Glp N2 and sgk-1(ok538) fed with UV-killed OP50 starting as 1-day old adults is plotted. (F) The sgk-1(ok538) mutant has a strong developmental delay, which is substantially weaker with sgk-1 RNAi knockdown. Thus, we used RNAi knockdown to assess the role of sgk-1 in pathogen resistance without developmental timing as a confounding factor. RNAi knockdown of cdc-25.1 and sgk-1 was performed serially. cdc-25.1 RNAi knockdown was performed maternally followed by egg lay to undiluted sgk-1 RNAi bacteria. RNAi knockdown of sgk-1 did not enhance pathogen resistance (logrank, p = 0.27). The efficacy of the induction of RNAi was confirmed by RNAi knockdown of daf-16, which enhanced susceptibility to pathogen (Table S1). Moreover, RNAi knockdown of sgk-1 has been reported to extend lifespan to 160% of control levels (Hertweck et al. 2004). (G) pdk-1(sa680) mutants were also not significantly different than N2 in resistance to PA14 (logrank test, p = 0.12). (H) The pathogen resistance of gain-of-function mutants in akt-1 and pdk-1 mirrored the loss-of-function mutants. akt-1(mg144) was sensitive compared to N2 (logrank p < 0.0001), whereas pdk-1(mg142) was not significantly different from N2 (logrank p = 0.6). (I) Relative mean survival on PA14 for experiments shown in Figure 2. (J) Relative mean lifespan on UV-killed OP50 for experiments shown in Figure 2. All experiments used cdc-25.1 RNAi Glp animals. Representative experiments shown. In panels C–E, representative lifespan and pathogen resistance assays were combined in the same plot to ease visual comparison. Complete supporting data are presented in Supplementary Table S1.