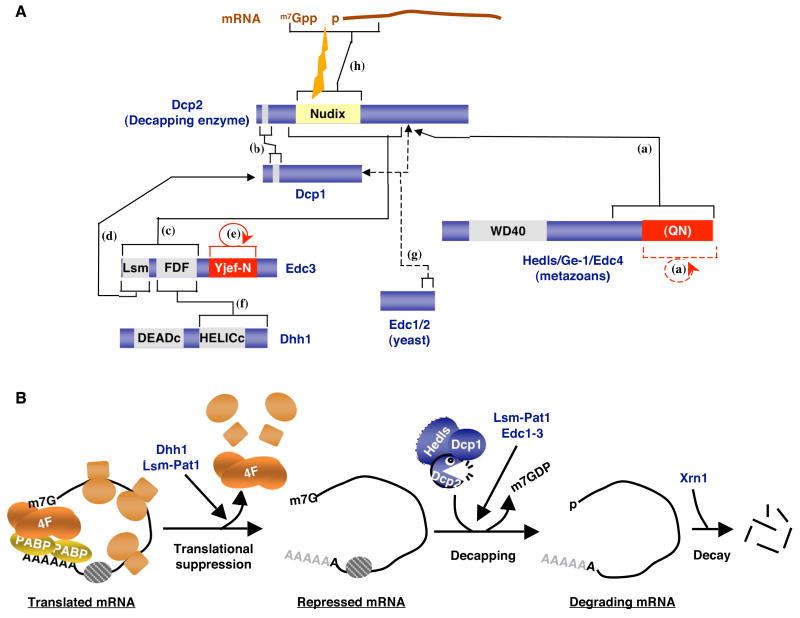
Figure 1. Multiple protein factors promote mRNA decapping.
(A) Interactions between the decapping enzyme Dcp2 and factors that stimulate decapping, Dcp1, Edc3, Dhh1 (Rck/P54, Me31B, CGH-1, Xp54), metazoan-specific Hedls and yeast-specific Edc1/2. Direct interactions between the decapping factors and between Dcp2 and mRNA are indicated by solid lines and brackets. The interaction between Edc1/2 and Dcp1/2 has been shown indirectly, through activation of decapping in vitro and is represented by dashed lines. Putative PB assembly domains are shown in red. Specific protein domains are indicated. (Q/N): Putative glutamine/asparagine-rich region of Hedls. The catalytic Nudix domain of Dcp2 is indicated in yellow. The indicated interactions are based on observations described in: (a): (Fenger-Gron et al., 2005; Xu et al., 2006; Yu et al., 2005), (b): (Deshmukh et al., 2008; She et al., 2006; She et al., 2008), (c): (Decker et al., 2007; Tritschler et al., 2007), (d): (Tritschler et al., 2007; Fenger-Gron et al., 2005), (e): (Decker et al., 2007; Tritschler et al., 2007), (f): (Decker et al., 2007; Tritschler et al., 2007), (g): (Dunckley et al., 2001; Schwartz et al. 2000), and (h): (Deshmukh et al., 2008; She et al., 2006; She et al., 2008). (B) mRNA decapping activation is thought to occur in two distinct steps. First, the cap-binding complex (eIF4F) must be removed from the mRNA, a process that renders the mRNA translationally inactive. This process is stimulated, at least in yeast, by Dhh1 and the Lsm-Pat1 complex. The core decapping complex, including Dcp2, Dcp1 and possibly in metazoans Hedls, can then access and remove the mRNA cap through a process that can be stimulated by Edc1–3. The decapped mRNA is then degraded by Xrn1.