Figure 6.
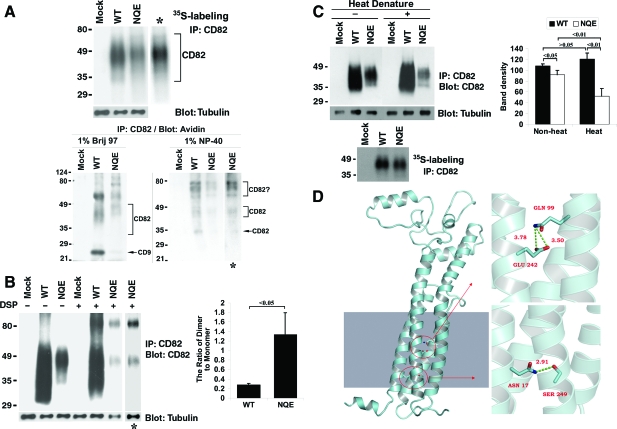
The TM polar residues are not essential for CD82 glycosylation and dimerization but are needed for its conformational stability. A: The NQE mutant is glycosylated. Top: Cells were labeled overnight with S35-methionine, lysed in 1% NP-40, and then immunoprecipitated with CD82 mAb TS82b. After SDS-PAGE, the immunoprecipitates were visualized by autoradiography. Bottom: Du145 transfectant (Mock, CD82 WT, and NQE mutant) cells were biotinylated at 4°C for 1 hour and lysed in either 1% Brij 97 or 1% NP-40. The lysates were co-immunoprecipitated with CD82 mAb M104. The precipitates were resolved in nonreducing SDS-PAGE followed by transfer to nitrocellulose membrane. The membrane was blotted with Extravidin, and the detected proteins were visualized by chemiluminescence. B: The NQE mutant forms dimer. Left: Du145 transfectant cells were cross-linked with DSP as described in Materials and Methods, treated with 1% NP-40, immunoprecipitated with CD82 mAb, and then immunoblotted with the same mAb. β1-tubulin in the same cell lysates was used as an internal loading control. The asterisks indicate the longer exposure of the NQE lanes. Right: The ratios of dimer to monomer were quantified based on the densities of dimer and monomer bands and are presented as the average ± SD of the results from 3 experiments. P < 0.05. C: The TM polar residues are needed for the conformational stability of CD82. Left: Du145-Mock, -CD82 wild-type, and -CD82 NQE transfectant cells in equal number were lysed with 1% Brij 97 lysis buffer. CD82 in the lysates was immunoprecipitated by its mAb TS82b and then treated with or without denature at 95°C for 5 minutes. After nonreducing SDS-PAGE separation and transferring to nitrocellulose membrane, CD82 proteins on the membrane were probed with its mAb TS82b, followed by the peroxidase-conjugated second Ab blot and chemiluminescence. β-tubulin from the same cell lysates, detected by Western blot, was used as an internal loading control. CD82 proteins immunoprecipitated from an equal number of S35-labeled Du145-Mock, -wild-type, and -NQE transfectant cells and revealed by autoradiography after SDS-PAGE separation were also included as loading control. Right: The CD82 bands were quantified with densitometry analysis, the band density of CD82 was normalized with the one of tubulin, and the average band densities of CD82 ± SD from three experiments were plotted as the histogram. P values are indicated in the histogram. D: The interactions of polar residues in homology model of CD82. An atomic model of CD82 was constructed with homology model followed by 5000-step energy minimization in AMBER, shown in the left section. The shaded area in the left mimics the membrane. The detailed interactions of polar residues N17, Q99, and E242 were displayed in enlarged images in the right part. The upper right image shows the locations of Q99 in TM3 and E242 in TM4, both of which point to inside TM helix bundle. The bottom right one represents the interaction between N17 in TM1 and S249 in TM4, which forms a hydrogen bond with a distance of 2.91Å.