Abstract
The yeast Sir1 protein's ability to bind and silence the cryptic mating-type locus HMRa requires a protein-protein interaction between Sir1 and the origin recognition complex (ORC). A domain within the C-terminal half of Sir1, the Sir1 ORC interaction region (Sir1OIR), and the conserved bromo-adjacent homology (BAH) domain within Orc1, the largest subunit of ORC, mediate this interaction. The structure of the Sir1OIR-Orc1BAH complex is known. Sir1OIR and Orc1BAH interacted with a high affinity in vitro, but the Sir1OIR did not inhibit Sir1-dependent silencing when overproduced in vivo, suggesting that other regions of Sir1 helped it bind HMRa. Comparisons of diverged Sir1 proteins revealed two highly conserved regions, N1 and N2, within Sir1's poorly characterized N-terminal half. An N-terminal portion of Sir1 (residues 27 to 149 [Sir127-149]) is similar in sequence to the Sir1OIR; homology modeling predicted a structure for Sir127-149 in which N1 formed a submodule similar to the known Orc1BAH-interacting surface on Sir1. Consistent with these findings, two-hybrid assays indicated that the Sir1 N terminus could interact with BAH domains. Amino acid substitutions within or near N1 or N2 reduced full-length Sir1's ability to bind and silence HMRa and to interact with Orc1BAH in a two-hybrid assay. Purified recombinant Sir1 formed a large protease-resistant structure within which the Sir1OIR domain was protected, and Orc1BAH bound Sir1OIR more efficiently than full-length Sir1 in vitro. Thus, the Sir1 N terminus exhibited both positive and negative roles in the formation of a Sir1-ORC silencing complex. This functional duality might contribute to Sir1's selectivity for silencer-bound ORCs in vivo.
Chromatin, the protein-DNA complex that comprises eukaryotic chromosomes, varies substantially with chromosomal position, and this structural heterogeneity is fundamental to genome function. A central question in chromosome biology concerns the mechanisms that establish this structural variation in the genome. Silencing of the cryptic mating-type locus HMRa in budding yeast is a powerful model for examining mechanisms that target and confine the formation of a specialized form of chromatin to specific regions of the genome (20, 46). Silencing is caused by the formation of “silent” chromatin that is analogous to metazoan heterochromatin, causing heritable, position-dependent transcriptional repression, delayed replication time, and inaccessibility of the chromosomal DNA to a variety of DNA-modifying enzymes (46).
Silent chromatin is targeted to HMRa by protein-DNA and protein-protein interactions that require the origin recognition complex (ORC) (20), the evolutionarily conserved multisubunit protein complex best known for its role in the initiation of eukaryotic DNA replication (2). ORC, along with additional sequence-specific DNA-binding proteins Rap1 and Abf1, binds to a small, ∼150-bp DNA sequence element called the HMR-E silencer. HMR-E is both necessary and sufficient to nucleate assembly of a silent chromatin domain that encompasses HMRa. Because of its role in DNA replication, ORC is essential for viability, as are the abundant multifunctional nuclear proteins Rap1 and Abf1. In contrast, silent chromatin and the four silent information regulator (SIR) proteins required for it are not essential. At HMR-E the silencer-binding proteins ORC, Rap1, and Abf1 come together to form a unique protein-DNA surface that can bind a complex of SIR proteins.
The SIR proteins play direct roles in the nucleation, assembly, and ultimate structure of silent chromatin at HMRa (23, 27, 41). A working model posits that Sir1 and Sir4 bind the silencer-binding proteins directly and stably enough to recruit the two other SIR proteins, Sir2 and Sir3, to the silencer (47). Once positioned at the silencer, Sir2, a NAD+-dependent histone deacetylase (16), removes acetyl groups from neighboring nucleosomes, which in turn enhances Sir3 binding to nucleosomes adjacent to HMR-E (7). As Sir2, Sir3, and Sir4 form a complex (14, 37, 45), this binding facilitates further Sir2-dependent deacetylation of nucleosomes that comprise HMRa until a stable silent chromatin structure is formed. In this model Sir1, unlike the three other Sir proteins, is not an essential structural component of silent chromatin. Instead, Sir1 targets the assembly of silent chromatin to HMRa by binding the HMR-E silencer. It is well established that this binding requires a direct and unique protein-protein interaction between Sir1 and ORC (4, 22, 30, 32, 55, 57).
A role for ORC in heterochromatin is conserved from yeast through metazoans (36, 49), and the Sir1-ORC interaction has served as a paradigm for understanding how ORC acquires locus-specific roles in chromatin structure (20). A minimal domain within Sir1, the Sir1 ORC interaction region (Sir1OIR), binds ORC through the N-terminal region of Orc1, the largest subunit of ORC (4). This region of Orc1 forms a bromo-adjacent homology (BAH) domain (3, 57). BAH domains are conserved among Orc1 orthologs (24) and are also found in a number of other chromatin-associated proteins (6, 26), suggesting that they have a fundamental role in chromatin structure (13, 42). Recent studies have provided high-resolution structural insights into the formation of the Sir1OIR-Orc1BAH complex (30, 32). In particular, one module within the Sir1OIR structure contains several amino acids that comprise a continuous surface that directly contacts a complementary surface on the Orc1BAH domain. Individual amino acid substitutions within this Sir1 silencer recognition-defective (SRD) module prevent Sir1 from binding Orc1BAH or ORC in vitro (4, 22) and from binding and silencing HMRa in vivo (21, 50).
In contrast to Orc1, Sir1 is only weakly conserved within the Saccharomyces genus. However, the SRD module is exceptional for its strong conservation among diverged Sir1 proteins (4), suggesting that Sir1's ability to bind ORC is one of its most constrained functions. In this study, we identified two other short regions within Sir1 that show high conservation between diverged Sir1 proteins. Amino acid substitutions within these regions produced mutant Sir1 proteins incapable of either silencing or binding HMRa in vivo or interacting with Orc1BAH in a two-hybrid assay. The latter observation was unexpected because the Sir1OIR is sufficient for a two-hybrid interaction with Orc1BAH (4), and it raised the possibility that the N-terminal region of Sir1 modulated the Orc1BAH-Sir1OIR interaction. Consistent with this possibility, in vitro experiments with purified recombinant proteins revealed that Sir1OIR interacted more efficiently than full-length Sir1 with Orc1BAH. Sir1 formed a single large protease-resistant domain that extended from the Sir1 N-terminal region and included the Sir1OIR. Based on these and other data, the N terminus of Sir1 had both positive and negative roles in forming a stable Sir1-ORC silencing complex.
MATERIALS AND METHODS
Yeast strains and plasmids.
Yeast strains and plasmids were constructed using standard yeast molecular genetics (28) and recombinant DNA techniques (35, 38, 48) and are listed in Tables 1 and 2, respectively.
TABLE 1.
Saccharomyces cerevisiae strains
| Straina | Genotype | Reference |
|---|---|---|
| JRY19 | MATahis4 leu2 trp1 ura3 | 22 |
| JRY2234 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 (W303-1A) | 54 |
| JRY3009 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 (W303-1B) | |
| CFY345 | JRY3009 HMR-SSa | 19 |
| CFY762 | JRY3009 HMR-SSasir1Δ::LEU2 | 22 |
| CFY932b | MATatrp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ sir2Δ::TRP1 | 33 |
| CFY770 | JRY3009 HMR-SSa (ACS GAL4 ABF1) sir1Δ::LEU2 | 22 |
| CFY1463 | JRY3009 HMR-SSaSIR1-HA3-kanMX6 | 4 |
| CFY1487 | JRY3009 HMR-SSasir1R493G-HA3-kanMX6 | 4 |
| CFY1535 | JRY3009 HMR-SSasir1n1-HA3-kanMX6 | |
| CFY1538 | JRY3009 HMR-SSasir1n2-HA3-kanMX6 | |
| CFY1583 | JRY3009 HMR-SSasir1NΔ-HA3-kanMX6 | |
| CFY1804 | JRY3009 HMR-SSasir3Δ::URA3 | |
| CFY2079 | JRY3009 HMR-SSasir3BAHΔ | |
| CFY2665 | JRY3009 HMR-SSasir1D36A-HA3-kanMX6 | |
| CFY2664 | JRY3009 HMR-SSasir1W38A-HA3-kanMX6 | |
| CFY2666 | JRY3009 HMR-SSasir1Q27A,N29A-HA3-kanMX6 |
All strains except JRY19 and CFY932 are isogenic derivatives of W303.
CFY932 is a sir2Δ::TRP1 derivative of the two-hybrid strain described in reference 33.
TABLE 2.
Plasmids
| Plasmid | Description | Reference |
|---|---|---|
| pTT65 | GAD-Orc1(5-228) | 55 |
| pCF413 | pADH1-GBD-SIR1 in pRS426 | 22 |
| pCF409 | pADH1-GBD-SIR1 in pRS416 | |
| pCF415 | pADH1-GBD-sir1V490D in pRS426 | 51 |
| pCF1520 | pADH1-GBD-sir1n1 in pRS426 | |
| pCF1522 | pADH1-GBD-sir1n2 in pRS426 | 22 |
| pCF721 | pADH1-GBD-SIR1(473-678) in pRS426 | |
| pCF1879 | pADH1-GBD-sir1D36A in pRS426 | |
| pCF1878 | pADH1-GBD-sir1W38A in pRS426 | |
| pCF1880 | pADH1-GBD-sir1Q27A,N29A in pRS426 | |
| pCF1881 | pADH1-NLS-SIR1OIR in pRS426 | |
| pCF1883 | pADH1 in pRS426 | |
| pCF1618 | pADH1-GAD-SIR3BAH | |
| pCF1861 | pADH1-SIR1(25-155)-GBD | |
| pCF91 | pMET3 in pRS313 | 18 |
| pCF1917 | pMET3-SIR1 in pRS313 | |
| pCF1916 | GST-ORC1(5-219) in pGEX-KG | 55 |
| pCF1748 | ORC1(5-219) in pET28b | 30 |
| pCF1586 | SIR1(480-611)C593A in pET28b | 30 |
| pCF1808 | SIR1(25-678)L26V in pET28b |
Measuring silencing.
The efficiency of HMRa silencing was determined by measuring a1 mRNA levels by RNA blot hybridization or by mating as described previously (8).
Immunoprecipitations, immunoblotting, and ChIP assays.
Three different antibodies were used as appropriate, as indicated in the figure legends. A Sir1 rabbit polyclonal antibody raised against purified Sir1OIR (32) (Harlan) or an antihemagglutinin (anti-HA) monoclonal antibody (12CA5 or 16B12) (Covance) was used to detect and/or immunoprecipitate Sir1 or Sir1-HA3. For Sir3 chromatin immunoprecipitation (ChIPs), a cocktail of anti-Sir3 monoclonal antibodies was used (8). This antibody was raised against full-length Sir3 purified from baculovirus-infected Sf9 cells as described previously (25) (Neoclone). Protein immunuprecipitations used antibodies that had been covalently cross-linked to protein A-Sepharose (Amersham Biosciences) using standard methods (29).
To ensure that equivalent amounts of protein were compared in protein immunoblots, Ponceau S staining of the filter and cross-reactivity between the antibodies and nonspecific (or specific, if appropriate) target proteins were used.
For ChIP experiments, 50-ml yeast cultures were grown to an optical density of 1.0 in yeast-peptone-dextrose or Casamino Acids medium. After formaldehyde cross-linking, quenching, and cell disruption, the resulting chromatin-containing supernatant was collected in a 2-ml Eppendorf tube and sonicated using a Branson 250 sonicator. The sonicated material was centrifuged to remove cellular debris, and the soluble supernatant (chromatin solution) was transferred to a new tube and incubated with 50 μl of the appropriate antibody-coupled Sepharose beads and 2 μl of sheared lambda DNA at 4°C overnight. Samples were centrifuged, and the supernatant (total sample) was transferred to an Eppendorf tube with 400 μl of TE-SDS (50 mM Tris-HCl [pH 8.1], 1% sodium dodecyl sulfate [SDS], 1 mM EDTA). The pellet containing the immunoprecipitated material was washed once in lysis buffer; once in lysis buffer plus 500 mM NaCl; once in 10 mM Tris-HCl (pH 8.1), 0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, and 1 mM EDTA; and once in TE. One hundred microliters of TE-SDS was added to the washed resin. Total and immunoprecipitated samples were incubated at 65°C overnight and briefly centrifuged, and the supernatant transferred to fresh Eppendorf tubes containing 5 μl of proteinase K (20 mg/ml) and incubated at 37°C for 1 h. DNA was purified, and specific fragments were detected by PCR with sequence-specific DNA primer pairs. PCR products were analyzed on a 1% agarose gel, stained with ethidium bromide, and quantified using the Epi Chemi II Dark Room system and LabWorks Analysis software (UVP Laboratory Products). Data are presented as the percentages of immunoprecipitated HMR-SSa and ADH4 DNAs recovered from the starting (total) sample. Primers used to detect HMR-SSa flanked the silencer. The HMR-SSa and ADH4 primers were as described previously (8).
Isothermal titration calorimetry.
Sir1OIR and Orc1BAH were purified as described previously (24) and dialyzed extensively in 25 mM phosphate buffer (pH 7.2)-200 mM NaCl. (Sir1OIR contained an amino acid substitution [C593A] to enhance solubility of the domain; this substitution did not affect the Sir1OIR-Orc1BAH interaction but facilitated crystallization of Sir1OIR and Sir1OIR-Orc1BAH complex [24].) Protein binding assays were performed on a Microcal VP-ITC isothermal calorimeter (Microcal, Amherst, MA); 145 μM Orc1BAH was injected into the sample cell containing 6.5 μM Sir1OIR at 23°C. The change in heat after each titration step was obtained by peak integration using Origin data analysis software provided by Microcal (OriginLab Corp., Northampton, MA). The binding constant (Ka) between these two domains was obtained by fitting the data to a single-site binding curve. The dissociation constant (Kd) was calculated as 1/Ka.
Homology modeling to predict the structure of the N-terminal region of Sir1.
The Sir1 protein sequence from residues 25 to 224 was submitted to the Robetta full-chain protein structure prediction server (www.robetta.org) (10-12, 34, 44). The server separated the peptide into two parts that were modeled separately. Part I included residues 25 to 153 and was modeled based on its similarity to the Sir1OIR. The confidence score for part I was 53, which is much higher than 3, the cutoff for good modeling. (The confidence score is related to the P value determined by the PSI-BLAST search and does not reflect the actual quality of the predicted structure, but high confidence means that there is a high probability that the program will produce predicted structures that are close to the actual structure.) In the 10 structures produced for part I, 4 were modeled using Sir1OIR structure from PDB file 1Z1A (Sir1OIR alone) chain a, and 6 were modeled using Sir1OIR structure from PDB file 1ZHI (Sir1OIR-Orc1BAH complex) chain b. All 10 structures were similar and closely resembled the Sir1OIR structure.
Protein expression and purification.
Sir1OIR and Orc1BAH were purified as described previously (24). For expression and purification of Sir1, DNA encoding Saccharomyces cerevisiae Sir1 residues 25 to 678 was cloned into the NcoI and HindIII sites of pET28b (Novagen) with a His6 epitope tag immediately following the last amino acid residue of Sir1, creating pET28b-Sir1-6×His. The codon for amino acid residue L26 of Sir1 was changed to a valine due to the requirement of a NcoI restriction site. For expression, Rosetta(DE3)pLys cells (Novagen) transformed with pET28b-Sir1-6×His were grown in LB medium supplemented with 30 μg/ml kanamycin and 15 μg/ml chloramphenicol to an A600 of ∼0.5. The culture was cooled to 16°C for inducing protein expression with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 18 h. Cells were harvested by centrifugation, frozen at −80°C, resuspended at 4°C in lysis buffer [50 mM phosphate (pH 7.2), 300 mM NaCl, 10% glycerol, and 1 mM tris(2-carboxyethyl)phosphine] and lysed with a French press. The cell lysate was cleared by centrifugation at 38,000 × g for 15 min and then loaded onto a HiPrep 16/10 SP FF column (Amersham) preequilibrated with lysis buffer. Proteins were eluted with a linear (0.3 M to 0.85 M) NaCl gradient. Fractions containing Sir1 were pooled and supplemented with 20 mM imidazole prior to incubation with Ni-agarose (Amersham) for 1 h. The Ni-agarose resin was packed into a column and washed extensively with lysis buffer. Sir1 was eluted with lysis buffer containing 500 mM imidazole and further purified on a Superdex200 PG column (Amersham) equilibrated in Superdex200 buffer (50 mM phosphate [pH 7.2], 500 mM NaCl, 10% glycerol, and 1 mM dithiothreitol).
Gel filtration analysis of Sir1-Orc1BAH and Sir1OIR-Orc1BAH complexes.
Purified Sir1 or Sir1OIR proteins (12 μM) were mixed with 20 μM of Orc1BAH in Superdex200 buffer and incubated on ice for 30 min, and then 1.2 ml of the mixture was fractionated over a Superdex200 PG column equilibrated with Superdex200 buffer into 3-ml fractions. Seven microliters of each fraction was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Limited proteolysis of Sir1.
Forty-five microliters of purified Sir1 (1 mg/ml) was mixed with 5 μl of 29-μg/ml trypsin in Superdex200 buffer at room temperature. After the indicated incubation times (see Fig. 7C), 8 μl was removed, mixed with Laemmli buffer, and boiled for analysis by SDS-PAGE.
FIG. 7.
Sir1 is less effective than Sir1OIR in interacting with the Orc1BAH in vitro. (A) Recombinant full-length Sir1 (amino acids 25 to 678) purified from E. coli bound a GST-Orc1BAH fusion protein. (B) Gel filtration was used to compare the strength of a Sir1OIR-Orc1BAH interaction (top) to that of a full-length Sir1-Orc1BAH interaction (bottom). Fractions that eluted from the gel filtration column were examined by SDS-PAGE. Gels were stained with Coomassie blue. The elution positions of molecular mass standards on the gel filtration column are indicated at the top of the gel. (C) Purified Sir1 was subjected to limited proteolysis by trypsin at room temperature. Aliquots of Sir1 reaction mixtures incubated in the absence of trypsin for 30 min (time zero) or in the presence of limiting amounts of trypsin for the indicated times were analyzed by SDS-PAGE. (D) The N termini of the three major trypsin fragments generated in panel C (numbered) were identified by Edman degradation sequencing. The C-terminal amino acid was deduced based on the fragment size and the positions of trypsin cleavage sites. The fragment indicated by an asterisk was full-length protein that remained after the 15-min limited trypsin digest. The N1, N2, SRD, and OIR are indicated in the diagram of Sir1 at the top. (E) A speculative model to explain how the N-terminal region of Sir1 could modulate the Sir1OIR-Orc1BAH interaction. Sir1 exists in two different conformations that can interconvert. One of these conformations (left) masks the Sir1OIR and prevents it from binding to the Orc1BAH domain. However, a weak and/or transient interaction between the Sir1 N terminus and a BAH domain (Sir1+BAH) could favor formation of the Sir1 conformation in which the Sir1OIR is fully exposed for a high-affinity interaction with Orc1BAH. In the two-hybrid and in vitro experiments, the BAH domain that weakly interacts with the Sir1 N terminus is Orc1BAH, but in vivo the putative BAH domain is unknown. Mutations that affect the N-terminal region of Sir1 might be defective in silencing and in the two-hybrid interaction with Orc1BAH in part because they disrupt a weak Sir1N-BAH interaction (sir1n+BAH), leading to a reduction in the levels of Sir1 in the conformation that presents the Sir1OIR for a high-affinity interaction with Orc1BAH. Overexpression of such a mutant might provide enough of the open-conformation form of Sir1 needed for silencing. Note that this abstract model attempts to address only the paradoxical Sir1-Orc1BAH interaction dynamics reported in this study. It is probable, based on measurements of silencing and Sir1 binding to the silencer, that the Sir1 N-terminal region plays a role in stabilizing Sir1 after it has established its interaction with ORC.
Edman degradation sequencing.
Limited proteolysis of Sir1 was performed with trypsin for 15 min as described above. The sample was then separated by 8% SDS-PAGE, transferred onto a polyvinylidene difluoride membrane, and visualized by Coomassie blue staining. Bands of interest were isolated and sequenced by the Tufts University Core Facility (www.tucf.org), Boston, MA.
RESULTS
Sir1OIR binds Orc1BAH efficiently in vitro but does not compete with wild-type Sir1 for silencing when overexpressed.
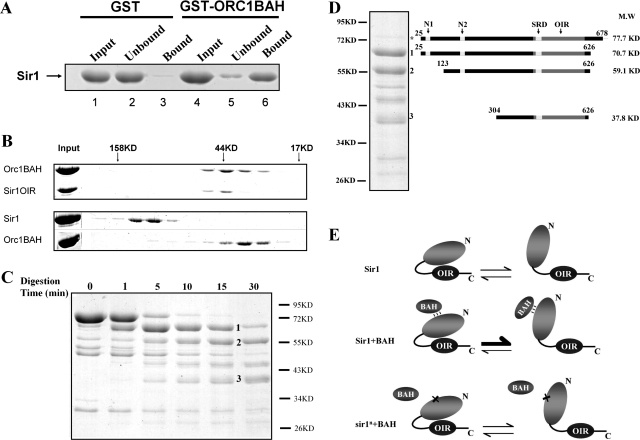
Sir1OIR is a small, 122-amino-acid domain within Sir1 that is sufficient to bind ORC and the Orc1BAH domain in vitro (4). The structure of the Orc1BAH-Sir1OIR complex reveals an extensive network of interactions (30, 32). To determine the affinity of the interaction between Sir1OIR and Orc1BAH, isothermal titration calorimetry experiments were performed with purified protein domains (Fig. 1A). These experiments indicated a Kd for the Sir1OIR-Orc1BAH complex of 0.20 μM in a solution containing 25 mM phosphate (pH 7.2) and 200 mM NaCl.
FIG. 1.
Sir1OIR binds Orc1BAH efficiently in vitro but does not compete with wild-type Sir1 for silencing when overexpressed. (A) Binding between Sir1OIR and Orc1BAH was measured by isothermal titration calorimetry. Sir1OIR and Orc1BAH were purified (30) and dialyzed extensively in 25 mM phosphate (pH 7.2)-200 mM NaCl. The protein domains were used at 145 μM for Orc1BAH and 6.5 μM for Sir1OIR. The Kd for the Sir1OIR-Orc1BAH interaction was calculated as 0.20 ± 0.03 μM. (B) Cell extracts from MATα SIR1 (CFY345) yeast were examined for Sir1 and Sir1OIR by protein immunoblotting with an anti-Sir1 antibody. One strain contained a vector carrying the ADH1 promoter (pCF1883); its only source of Sir1 was from the SIR1 locus (SIR1). The second strain was identical except that it contained a plasmid in which the ADH1 promoter drove expression of SIR1OIR fused to an NLS (SIR1OIR) (pCF1881). The extracts were mixed prior to SDS-PAGE at the indicated cell equivalent amounts, expressed in A600 units. The rabbit polyclonal anti-Sir1 used in this experiment was raised against Sir1OIR purified from E. coli. The Sir1OIR-containing-extract was diluted to 0.001 A600 unit to achieve signals comparable to that for 0.4 A600 unit of Sir1 extract (lane 4). The level of native Sir1 in each lane served as internal loading control; in addition, the filter was also stained with Ponceau S prior to immunoblotting to determine levels of protein loading and transfer that would allow interpretation. The Epi Chemi II Dark Room system and LabWorks Analysis software (UVP Laboratory Products) were used for quantification. (C) pADH1 (pCF1883)- or pADH1-NLS-SIR1OIR (pADH1-OIR; pCF1881)-containing plasmids were transformed into MATα HMR-SSa cells that were either SIR1 (CFY345) or sir1Δ (CFY762). Silencing of HMR-SSa was measured in semiquantitative mating assays in which 10-fold serial dilutions of cells being tested were mixed with an excess of MATa cells (CFY616) and plated to selective agar medium (Mating) that allowed only diploid cells to grow. The MATα cells used in the mating assay were simultaneously examined for viability on nonselective agar medium (Growth) to ensure that equivalent numbers of cells were used. (D) Established and potential functional regions of Sir1. The 122-amino-acid OIR binds the Orc1BAH domain, and its structure has been solved (30, 32). The SRD region is conserved between the diverged Sir1 proteins from S. castellii and S. cerevisiae and forms the surface on the Sir1OIR that is involved in direct contacts with Orc1BAH (30, 32). The Sir4 binding region is sufficient to produce a weak two-hybrid interaction with Sir4 (4, 55). The N1 and N2 regions are also highly conserved between these S. castellii and S. cerevisiae Sir1 proteins. A protein BLAST search with the Sir1OIR identifies a region within the Sir1 N terminus as 27% identical and 47% similar (OIR-similar). (E) Protein sequence alignment of Sir1 proteins from S. cerevisiae (top sequence) and S. castellii from amino acid 489 to 608 of S. cerevisiae Sir1. Invariant regions within the SRD module are highlighted. (F) Protein sequence alignment of Sir1 proteins from S. cerevisiae (top sequence) and S. castellii from amino acid 28 to 193 of S. cerevisiae Sir1. The two regions of amino acid identity, N1 and N2, are highlighted. The alignment used here was obtained from the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org/) using the Fungal Alignment option under the Comparison Resources toolbar.
If the Sir1OIR-Orc1BAH interaction was the major force for Sir1 binding to HMRa, then Sir1OIR, when overexpressed to sufficiently high levels in vivo, might compete with native Sir1 for binding to the silencer-bound ORC. Such competition would be predicted to have a dominant negative effect on silencing. To test this possibility, the Sir1OIR was fused to a nuclear localization signal (NLS) and expressed from the ADH1 promoter (pADH1) contained on a 2μm plasmid. In a separate experiment, expression of a green fluorescent protein-NLS-Sir1OIR fusion protein indicated that the NLS-Sir1OIR was sufficient for efficient nuclear localization of the protein (Z. Hou, unpublished data). To determine the level of overproduction of NLS-Sir1OIR relative to native Sir1, a fixed amount of cell extract from cells expressing native SIR1 was mixed with defined dilutions of extracts from isogenic cells expressing the pADH1-NLS-Sir1OIR. The resultant mixtures were analyzed in protein immunoblots with anti-Sir1 antibody (Fig. 1B). Native Sir1 appeared as a doublet in these experiments, and both bands were used for quantification (Epi Chemi II Dark Room system and LabWorks Analysis software [UVP Laboratory Products]). The NLS-Sir1OIR was expressed at levels approximately 400-fold greater than native Sir1 (Fig. 1B, lane 4) (full-length Sir1 and Sir1OIR produced nearly equivalent immunoblot signals when a mixture containing 0.4 A600 equivalents of SIR1 cells and 0.001 A600 equivalents of overexpressing-NLS-Sir1OIR cells was examined.) Assuming that the Sir1OIR could show some selectivity for silencer-ORCs these levels of Sir1OIR should be sufficient to compete native Sir1 from HMRa.
To determine whether overexpressed Sir1OIR competed with Sir1, the pADH1-NLS-Sir1OIR-containing 2μm plasmid (Fig. 1C, pADH1-OIR) or the corresponding empty plasmid (Fig. 1C, pADH1) were transformed into MATα HMR-SSa SIR1 cells. As a reference, the same plasmids were transformed into MATα HMR-SSa sir1Δ cells. HMR-SS is an engineered version of the HMR-E silencer that sensitizes HMRa silencing to Sir1 and ORC function and has been used extensively to dissect these proteins' roles at HMRa (19, 39, 43). pADH1-NLS-Sir1OIR did not inhibit silencing in SIR1 cells, nor was it able to provide for wild-type levels of SIR1 function in sir1Δ cells (Fig. 1C). Unexpectedly, however, pADH1-NLS-Sir1OIR was able to improve silencing in sir1Δ cells by about 10-fold, suggesting that this small domain alone could perform some of the silencing functions of SIR1. Regardless, these experiments established that Sir1OIR, although sufficient to bind the Orc1BAH domain with a high affinity in vitro (Fig. 1A), could not effectively compete with native Sir1 for silencing. Thus, mechanisms in addition to the Sir1OIR-Orc1BAH interaction must contribute to Sir1's selective and/or stable association with a silencer in vivo (4).
S. castellii and S. cerevisiae Sir1 proteins share only three short clusters of contiguous amino acid identity.
The regions of Sir1 known to be important for silencing reside within the C-terminal half of the 678-amino-acid Sir1 protein (Fig. 1D, OIR and Sir4 binding regions) (4). To identify other important regions of Sir1, we exploited the rapid divergence of SIR1 among closely related yeast species, since highly conserved regions between otherwise diverged proteins can help identify functionally relevant domains. For this purpose we examined Saccharomyces castellii, which contains three genes with similarity to S. cerevisiae SIR1 that have diverged enough to be potentially useful. Neither synteny nor phylogenetic methods could identify conclusively the SIR1 ortholog in S. castellii among these three diverged SIR1 genes. However, we chose to use an alignment of S. cerevisiae Sir1 with the S. castellii Sir1 on contig 561 because this particular S. castellii Sir1 contains a functional OIR that interacts with S. castellii Orc1 in a two-hybrid assay (4).
This sequence alignment showed that the Sir1 proteins from S. cerevisiae and S. castellii contig 561 share little amino acid identity (22% identity and 43% similarity; Fig. 1E and F show key portions of this alignment). However, studies of S. cerevisiae SIR1 that have defined Sir1 functional and structural domains (Fig. 1D) increased our confidence in the significance of this alignment. In particular, amino acids within the SRD module (Y489 to A505) that forms the ORC interaction surface of Sir1 are notable because they comprise a cluster of two invariant stretches of amino acids (S491 to L495 and F500 to E506) shared between these Sir1 proteins (4) (Fig. 1E). Thus, the SRD module is one of the most conserved regions between these diverged proteins. We reasoned that other regions with similar levels of amino acid identity might be important. Only two other regions between these Sir1 proteins are as strongly conserved as the region that comprises the SRD module (Fig. 1F). These regions, termed N1 (I35 to D41) and N2 (Q186 to L193), reside within the N-terminal half of Sir1, for which limited functional data are available (13, 17, 52).
N1 and N2 are required for Sir1 to silence and bind HMR.
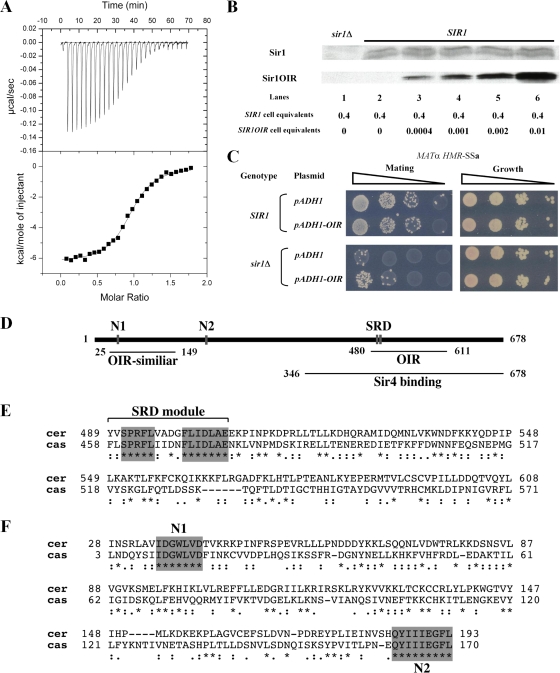
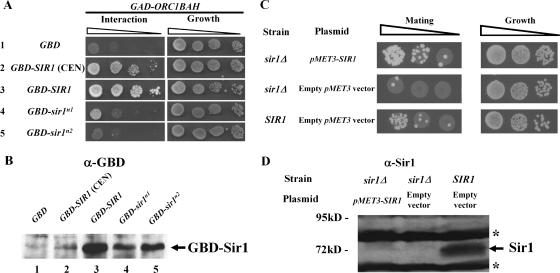
To test whether N1 and N2 contributed to SIR1 function, chromosomal SIR1 was replaced with relevant sir1 mutant alleles in MATα HMR-SSa cells. Wild-type and mutant Sir1 proteins were expressed from the chromosomal SIR1 locus with a C-terminal HA3 tag. Three different N-terminal mutants were analyzed: an N1 mutant (35-IDGWLVD-41 changed to 35-IAAALVA-41, referred to as sir1n1), an N2 mutant (185-QYIIIEGFL-193 changed to 185-QAAAAEGFL-193, referred to as sir1n2), and a mutant that lacks Sir1 codons 2 to 345, referred to as sir1NΔ. The mutations were designed to destroy any potential function of N1 or N2. The previously characterized sir1R493G allele was analyzed in parallel. This SRD allele produces a mutant sir1 protein defective in binding ORC (21, 22). The sir1R493G allele silences HMR-SSa about twofold more effectively than sir1Δ (mating efficiency of ∼5 × 10−3 [22]), and sir1Δ silences HMR-SSa over 3 orders of magnitude more effectively than sir2Δ, sir3Δ, or sir4Δ mutations, which abolish silencing (mating efficiencies of 1 × 10−6). Analysis of Sir1-HA3 levels indicated that each mutant version of Sir1 protein was expressed (Fig. 2A).
FIG. 2.
N1 and N2 are required for Sir1 to silence and bind HMR. (A) HA-tagged SIR1 alleles were integrated at the SIR1 chromosomal locus, and Sir1-HA3-tagged proteins were detected in protein immunoblots with an anti-HA antibody. * and ** indicate non-Sir1 proteins recognized by anti-HA. Except for the sir1 allele, the yeast cells used in this analysis were isogenic to a MATα HMR-SSa SIR1 strain that was also examined as a control (no tag). (B) RNA blot hybridization of a1 mRNA and SCR1 RNA from yeast cells described in panel A. SCR1 RNA served as a loading control. (C) Quantitative mating assays are performed by measuring the number of cells in a population of viable cells that can mate (and hence silence). The ratio of mating-competent cells to total number of cells is indicated on the y axis (which is shown in log scale) for each strain indicated. (D) Anti-Sir3-directed ChIP experiments. The averages and standard deviations from three independent experiments are shown. ChIPs were performed on a sir3Δ strain (CFY1804) to demonstrate the specificity of the anti-Sir3 antibody. HMR-SSa was detected with HMR-E silencer-specific primers. ADH4 served as a non-Sir1-dependent immunoprecipitation control. Primers used to detect HMR-SSa and ADH4 have been described previously (8). The percentage of HMR-SSa or ADH4 immunoprecipitated out of total starting DNA for each yeast strain is indicated with gray or black bars, respectively. (E) Anti-Sir1 directed ChIP experiments.
Silencing was measured in these yeast cells to compare the contributions of the N1, N2, and SRD regions to silencing of HMRa. Direct measurements of a1 mRNA by RNA blot hybridization revealed that these sir1 alleles reduced HMRa silencing similarly (Fig. 2B). As a second measure of silencing, these cells were assayed for their ability to mate (Fig. 2C). Simultaneous expression of a1 genes from HMRa and α genes from MAT generates a nonmating phenotype. Quantitative mating assays provided corroborating evidence that the N1 and N2 regions contributed to silencing. However, the sir1NΔ allele provided ∼10-fold better silencing than any of the other mutant alleles based on these assays, which is consistent with qualitative patch mating assays (J. R. Danzer, unpublished data). Nevertheless, sir1NΔ was defective compared to wild-type SIR1. Thus, the N-terminal portion of Sir1 was important for silencing.
A sir1Δ mutation reduces the level of Sir2, -3, and -4 proteins that bind the silencer (47). To address whether the silencing defects of sir1n1 and sir1n2 were caused by defects in Sir2-Sir24 recruitment, we performed ChIP experiments with an anti-Sir3 antibody. Sir3 binding is a good measure of Sir2-Sir4 complex formation at HMRa (46). Sir3 bound HMR-SSa efficiently and specifically in wild-type SIR1 cells but not in any of the sir1 mutant cells (Fig. 2D). Thus, regions within the Sir1 N terminus were required for Sir3, and by inference the Sir2-Sir4 protein complex, to bind HMRa.
Sir1 N1 and N2 could be important for recruiting Sir3 to the silencer by influencing the binding of Sir1 itself to the silencer. Alternatively, sir1n1 or sir1n2 mutant proteins could bind the silencer but be incapable of a subsequent step required to recruit Sir2-Sir4 to HMR-SSa. To distinguish between these possibilities, Sir1-directed ChIP experiments were performed (Fig. 2E). These experiments revealed that the mutant sir1n1 and sir1n2 proteins, like the reference sir1R493G protein (21, 50), failed to bind HMR-SSa. Thus, N1 and N2 were required for Sir1 to bind HMR-SSa in vivo.
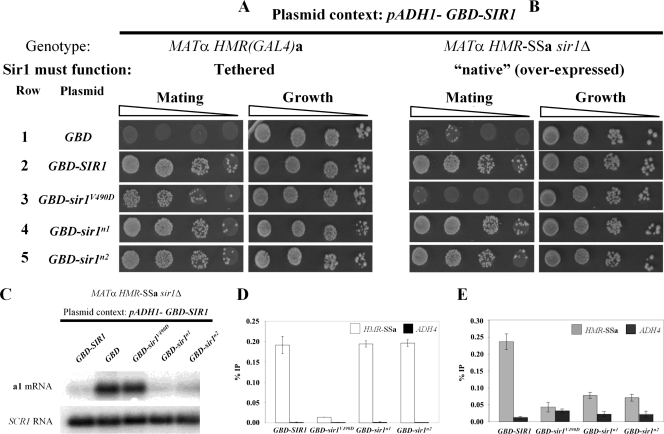
N1 and N2 are dispensable for HMRa silencing when Sir1 is tethered to HMRa or overproduced.
A fusion protein consisting of the Gal4 DNA binding domain (GBD) and Sir1 expressed under the control of the ADH1 promoter can silence an HMRa locus that contains a Gal4 DNA binding site within a mutant HMR-E silencer incapable of silencing by the native mechanism (9) (Fig. 3A, rows 1 and 2). Sir1-tethered silencing bypasses the requirement of chromosomal SIR1 and ORC in silencing but still requires the three other SIR genes (9, 18). Importantly, GBD-SIR1 can complement a sir1Δ mutation and bind HMR-E by the normal Sir1 mechanisms (e.g., via a Sir1-ORC interaction) (9, 22) (Fig. 3B, rows 1 and 2). In contrast to wild-type GBD-SIR1, a GBD-sir1srd allele (V490D) functions reasonably well in the tethered assay (22) (Fig. 3A, rows 1 and 3) but fails to complement a sir1Δ mutation in MATα HMR-SSa sir1Δ cells (22) (Fig. 3B, rows 1 and 3). This phenotype is caused by a defect in the ORC interaction surface of Sir1 that prevents formation of the Sir1-ORC complex (30, 32).
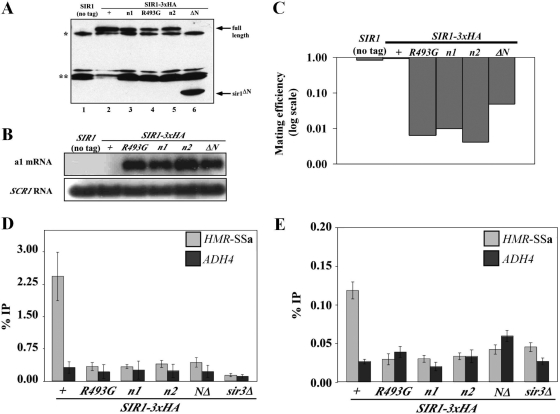
FIG. 3.
N1 and N2 are dispensable for HMRa silencing when Sir1 is tethered to HMRa or overexpressed. (A and B) 2μm plasmids containing the ADH1 promoter driving GBD (pCF394), GBD-SIR1 (pCF413), GBD-sir1V490D (pCF415), GBD-sir1n1 (pCF1520), and GBD-sir1n2 (pCF1522) fusions were transformed into MATα HMR-(GAL4)a sir1Δ (CFY770) cells (A) or MATα HMR-SSa sir1Δ (CFY762) cells (B). The transformed cells were assayed for silencing of the HMRa locus as determined by their ability to mate with an excess of MATa cells (CFY616). (C to E) The cells analyzed in panel B were analyzed for a1 mRNA by RNA blot hybridization (C), Sir3 binding to HMR-SSa by anti-Sir3-directed ChIPs (D), and Sir1 binding to HMR-SSa by anti-Sir1-directed ChIPs (E). Error bars indicate standard deviations.
The data presented thus far (Fig. 2) show that the sir1n1 and sir1n2 alleles caused phenotypes indistinguishable from those caused by an SRD allele, sir1R493G: all three alleles produced mutant proteins that failed to bind HMR-SSa, recruit Sir3 to HMRa, or silence transcription of the a1 gene. However, SRD alleles that affected the N-terminal coding region of SIR1 were never identified (22). Therefore, we assessed the ability of the sir1n1 and sir1n2 alleles to support HMRa silencing as GBD-SIR1 fusions in MATα HMR-(GAL4)a cells (tethered SIR1 silencing) (Fig. 3A) and MATα HMR-SSa sir1Δ cells (“native” [overexpressed] SIR1 silencing) (Fig. 3B).
These experiments revealed a striking difference between the sir1n and sir1srd alleles and explained why mutations in the N1 and N2 coding regions were not recovered in the original screen for SRD alleles (22). In contrast to GBD-sir1V490D, the GBD-sir1n1 and GBD-sir1n2 mutant proteins rescued silencing in both MATa HMR-(GAL4)a cells and MATα HMR-SSa sir1Δ cells. Thus, overproduced GBD-sir1n1 and GBD-sir1n2 could function by the native Sir1 mechanisms even though chromosomally produced sir1n1 and sir1n2 could not (Fig. 2). This observation was confirmed in a different context; sir1n1-3×HA or sir1n2-HA3, but not sir1V490D-HA3, could rescue silencing in MATα HMR-SSa sir1Δ cells when overproduced (J. R. Danzer, unpublished data). In addition, these observations were consistent with the findings of an earlier study that high-copy versions of SIR1 alleles containing mutations in an OIR-similar region of the Sir1 N terminus had no effect on normal silencing (13). Thus, in contrast to the SRD region, the Sir1 N-terminal regions N1 and N2 were not essential for silencing provided that enough Sir1 protein was made.
As an independent assessment, RNA blot hybridizations of a1 mRNA were performed (Fig. 3C). This experiment made it clear that the pADH1-GBD-sir1n alleles were as effective as wild-type SIR1 in silencing transcription of the a1 gene at HMRa (Fig. 3C). Therefore, a simple prediction was that these alleles would also be sufficient to restore Sir3 and Sir1 binding to HMR-SSa. To test this prediction, we performed Sir3- and Sir1-directed ChIPs (Fig. 3D and E).
The pADH1-GBD-sir1n alleles, but not pADH1-GBD-sir1V490D, restored wild-type levels of Sir3 binding to HMR-SSa (Fig. 3D), consistent with the ability of the sir1n alleles but not sir1srd to silence HMR-SSa when overproduced (Fig. 3B and C). In addition, pADH1-GBD-sir1n1 and pADH-GBD-sir1n2 but not pADH-GBD-sir1V490D partially restored Sir1 binding to HMR-SSa (Fig. 3E). Neither pADH1-GBD-sir1n1 nor pADH-GBD-sir1n2 produced as much fusion protein as wild-type pADH1-GBD-SIR1 (Fig. 4B), and this fact may explain why they produced less robust Sir1 binding to HMR-SSa than did wild-type pADH1-GBD-SIR1 (Fig. 3E). Nevertheless, the N1 and N2 regions, in contrast to the SRD region, were not absolutely essential for Sir1's association with HMR-SSa.
FIG. 4.
N1 and N2 contribute to the two-hybrid interaction between Sir1 and Orc1-BAH. (A) Two-hybrid interactions that controlled transcription of a HIS3 reporter gene were assessed as growth of cells on agar medium lacking histidine. A sir2Δ mutation was generated in the two-hybrid strain (CFY932) to avoid SIR-dependent silencing affecting the two-hybrid signal. To ensure that equal numbers of cells were evaluated in each experiment, all cells were also plated to synthetic complete agar medium. (B) The steady-state levels of the relevant GBD-Sir1 fusions were determined by anti-GBD (Babco) protein immunoblotting. To ensure that equivalent amounts of crude protein were assessed per lane, the filter was stained with Ponceau S prior to immunoblotting. (C) Mating was assessed for sir1Δ and SIR1 cells that were MATα HMR-SSa and carried a HIS3 CEN plasmid (pRS313) with pMET3-SIR1 (pCF1917) or with pMET3 (pCF91). The cells were grown in the presence of 2 mM methionine, which represses the MET3 promoter. (D) Levels of Sir1 expressed in the cells in panel C were assessed by a protein immunoblot with anti-Sir1OIR. To ensure that equivalent amounts of crude protein were assessed per lane, the filter was stained with Ponceau S prior to immunoblotting. The bands marked with an asterisk arise from nonspecific interactions between the antibody and proteins present in the extract and served as additional loading controls in this experiment.
N1 and N2 contribute to the two-hybrid interaction between Sir1 and Orc1BAH.
The aforementioned data provided evidence that N1 and N2 helped stabilize Sir1's binding to the silencer, but they did not address the mechanism of this stabilization. The following experiments (see Fig. 4 to 7) attempted to address possible mechanisms.
A characteristic of a sir1srd mutant protein is its inability to interact with ORC. This defect can be measured in a two-hybrid assay between Sir1 and Orc1BAH (4, 22). Although full-length GBD-SIR1 can interact with a Gal4 activation domain (GAD)-Orc1BAH fusion protein (Fig. 4A, rows 1 to 3), the N-terminal half of Sir1 is completely dispensable for the Sir1-Orc1BAH interaction (4, 55). These data make sense since the Sir1OIR is located in the C-terminal half of Sir1 (4, 30). N1 and N2 are in the N-terminal third of Sir1, far removed from the Sir1OIR that is sufficient to bind ORC. Nevertheless, given ORC's central role in recruiting Sir1 to HMRa, we tested whether N1 and N2 were required for full-length Sir1's two-hybrid interaction with Orc1BAH (Fig. 4A). Unexpectedly, these experiments provided evidence that N1 and N2 contributed to the Sir1-Orc1BAH two-hybrid interaction in the context of full-length Sir1 (Fig. 4A, rows 4 and 5).
We were concerned that reduced levels of GBD-sir1n1 and GBD-sir1n2 compared to wild-type GBD-Sir1 might explain these data, because both mutants produced less fusion protein than the wild type (Fig. 4B, lanes 3 to 5). Therefore, to test whether reducing wild-type GBD-Sir1 levels was sufficient to abolish the two-hybrid Sir1-Orc1BAH interaction, pADH1-GBD-SIR1 was expressed from a centromere-containing plasmid (Fig. 4B, lane 2). The levels of wild-type GBD-Sir1 were reduced substantially by expressing this fusion protein from a CEN plasmid, below the levels produced by either mutant (Fig. 4B, compare lane 2 to lanes 4 and 5). Nevertheless a robust two-hybrid interaction was produced (Fig. 4A, row 2). Thus, reduced levels of GBD-sir1n1 and GBD-sir1n2 were insufficient to account for their Orc1BAH interaction defects.
This experiment suggested that reductions in GBD-Sir1 levels did not dramatically affect the two-hybrid interaction signal produced between Sir1 and Orc1BAH, but we remained concerned that somewhat reduced levels of the Sir1 mutant proteins relative to the wild type expressed from our integrated alleles (Fig. 2A) might account for their silencing defects. Specifically sir1n1 and sir1n2, similar to sir1srd, were somewhat less abundant than wild-type Sir1 when expressed from the chromosomal SIR1 locus (Fig. 2A). However, unlike the sir1srd mutation, both sir1n1 and sir1n2 could silence when overexpressed (Fig. 3), raising the concern that they were silencing defective merely because they provided insufficient levels of Sir1 for silencing. Therefore, we attempted to reduce Sir1 levels artificially and measure the impact on silencing. MATα HMR-SSa cells that were either sir1Δ or SIR1 were transformed with a plasmid containing pMET3-SIR1 or simply the pMET3 promoter and grown in the presence of methionine. Methionine represses transcription by pMET3, and this promoter has been used to control the expression of a Gal4-Sir1 fusion (18). Under these conditions, sir1Δ cells containing pMET3-SIR1 silenced as efficiently as SIR1 cells containing a pMET3 vector (Fig. 4C). To measure Sir1 protein levels expressed in these cells, crude extracts were separated by SDS-PAGE and Sir1 was detected by protein immunoblotting (Fig. 4D). Sir1 levels were barely detectable from pMET3-SIR1 cells and were clearly below the level of Sir1 produced from the chromosomal locus. These data provided evidence that wild-type Sir1 levels could be reduced substantially without causing a silencing defect. Thus, reduced levels of Sir1 protein were unlikely to be the explanation for sir1n allele silencing defects.
Homology modeling of the Sir1 N terminus to guide the design of new sir1 alleles.
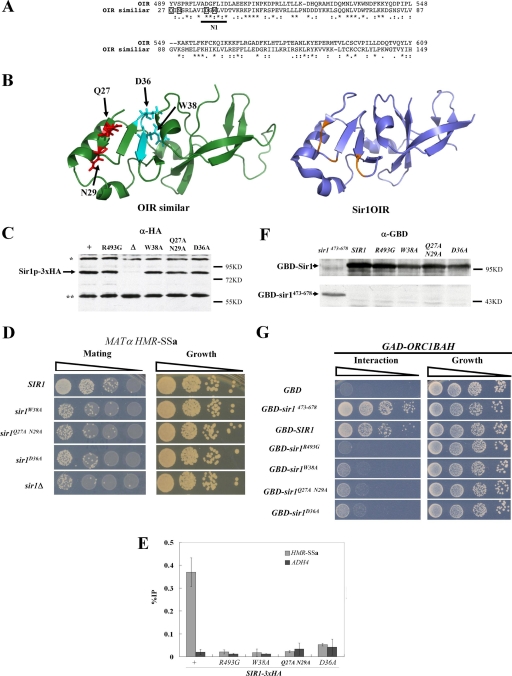
sir1n1 and sir1n2 were designed as multiple alanine substitution mutations to obliterate the potential function of highly conserved N-terminal stretches of Sir1 (Fig. 2). However, if N1 or N2 was required for a protein-protein interaction that was underlying the phenotypes caused by sir1n1 and sir1n2, we would expect that less drastic substitutions should produce similar phenotypes. Therefore, new alleles affecting N1 were designed based on sequence similarity between the Sir1OIR and the Sir1 N-terminal region (13), with a goal of avoiding substitutions that could cause major structural defects (Fig. 5A). Specifically, the solved structure of Sir1OIR was used by the Robetta full-chain protein structure prediction server (www.robetta.org) in a homology-based modeling algorithm to predict the structure of the Sir1 N-terminal region from amino acid 27 to 149 (Fig. 5B) (10-12, 34, 44). The predicted structures were used to guide the design of three new sir1 alleles (Fig. 5B). The sequence similarity (27% identity and 47% similarity) between these Sir1 regions allowed 10 high-confidence predicted structures of the Sir127-149 region to be generated. Each was similar to the one shown in Fig. 5B.
FIG. 5.
Homology modeling of the Sir1 N terminus to guide the design of new sir1 alleles. (A) Sequence alignment showing similarity between the Sir1OIR (top) and the N-terminal region of Sir1, originally presented in reference 13. This N-terminal region contains the phylogenetically conserved N1 region (Fig. 1), which is underlined. The amino acids within the Sir1 N-terminal region that were changed to alanines to generate the three different sir1 alleles for these experiments are boxed. (B) Modeling of the Sir1 N-terminal structure from amino acid 27 to 149 was performed using the Robetta full-chain protein structure prediction server (www.robetta.org) based on similarity of the N terminus to the Sir1OIR shown in panel A. Ten predicted structures were generated, all of which were similar to the structure shown on the left. The structure of the Sir1OIR is shown on the right for comparison. The ribbon diagrams were made using PYMOL (15). The representative structure of the Sir1-N terminus used to guide mutational analyses is shown, as well as stick diagrams of the amino acids that were targeted for substitution. (C) Three new sir1 alleles were integrated and tagged with the HA3 epitope at the native SIR1 locus. The levels of HA-tagged Sir1 protein generated by each allele were determined by anti-HA protein immunoblotting. * and ** indicate non-Sir1 cross-reacting proteins. (D) The ability of these alleles to provide for SIR1 function in silencing HMR-SSa was assessed by mating. (E) Sir1-HA3 binding to HMR-SSa was determined by anti-HA-directed ChIPs. Error bars indicate standard deviations. (F) Levels of GBD fusion proteins were assessed with anti-Gal4 DBD protein immunoblots. (G) Sir1-Orc1BAH two-hybrid interactions using the protein fusions in panel F were assessed as described for Fig. 4A for the GBD-Sir1 fusions indicated.
Three new mutant alleles affecting the N1 region were made. First, the SIR1 codon for aspartate 36 was changed to an alanine to create sir1D36A. Second, codon 38 for tryptophan was changed to an alanine to create sir1W38A. Third, codons 27 and 29 were changed to create sir1Q27A,N29A. This third allele was generated because the corresponding affected residues within the Sir1OIR are positioned directly on the surface that contacts the Orc1BAH in the Sir1OIR-Orc1BAH complex. The structural model predicts that these amino acid substitutions would have minimal effects on the structural integrity of the domain (Fig. 5B).
The new sir1 alleles were integrated as HA-tagged versions of SIR1 at the native SIR1 locus. Levels of Sir1 protein, HMR-SSa silencing, and Sir1 binding to HMR-SSa were assessed (Fig. 5C to E). The mutant proteins were expressed (Fig. 5C) but showed defects in silencing and binding to HMR-SSa (Fig. 5D and E). Thus, multiple independent alleles affecting the N1 region of Sir1 produced silencing phenotypes identical to those produced by sir1n1 and sir1n2.
The most difficult phenotype to explain with respect to the sir1n alleles was their inability to interact with a GAD-Orc1BAH fusion in a two-hybrid assay (Fig. 4). Therefore each of the new alleles was expressed as pADH1-GBD-SIR1 fusions. GBD-sir1W38A, GBD-sir1Q27A,N29A, and GBD-sir1D36A produced fusion proteins that failed to produce a robust two-hybrid interaction with GAD-Orc1BAH (Fig. 5F and G). In addition, this experiment assessed the GBD-sir1473-678 fusion, which lacks the entire N-terminal 472 amino acids of Sir1. This shorter fusion produced less protein than wild-type GBD-SIR1 yet interacted more robustly with GAD-Orc1BAH (Fig. 5G). The two-hybrid cells used in these experiments were sir2Δ to avoid the effects of silencing itself on the two-hybrid interaction signals. These data provided additional evidence that amino acids within the N1 region of Sir1 contributed to its two-hybrid interaction with Orc1BAH even though the entire N-terminal region of Sir1 was dispensable, and in fact somewhat inhibitory, for this interaction.
Sir3BAH does not mediate the roles of N1 and N2 in Sir1's binding to the silencer.
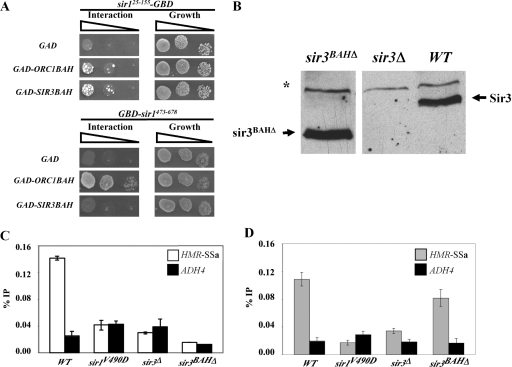
The Sir1 N-terminal region that includes N1 is similar to the Sir1OIR that binds the Orc1BAH domain (Fig. 5A and B). Therefore, it was reasonable to propose that the Sir1 N-terminal region might also interact with a BAH domain and that such an interaction might contribute to the roles of N1 and/or N2 in Sir1's stable association with HMRa. Indeed, an interpretation of the two-hybrid experiments in Fig. 4 and 5 was that the Sir1 N terminus had some affinity for Orc1BAH. Therefore, we tested whether a Sir1 N-terminal region from amino acid 25 to 155 that included N1 (Sir125-155-GBD) could interact with Orc1BAH in a two-hybrid assay. An extremely weak but reproducible two-hybrid interaction between Sir125-155-GBD and GAD-Orc1BAH was observed (Fig. 6A).
FIG. 6.
Sir3BAH does not mediate the roles of N1 and N2 in Sir1's binding to the silencer. (A) Two-hybrid interactions between sir125-155-GBD and GAD-Orc1BAH or GAD-Sir3BAH. The interaction of GBD-sir1473-678 with these same fusion proteins was also assessed for comparison. (B) A mutant allele of SIR3 lacking the coding region for the N-terminal Sir3 BAH domain (sir3BAHΔ) was integrated at the native SIR3 locus (CFY2079). Protein levels of both wild-type (WT) (CFY345) and mutant (CFY2079) Sir3 were determined by anti-Sir3 protein immunoblotting. A sir3Δ strain (CFY1804) was included as a negative control. (C and D) The cells from panel B and sir1V490D cells were examined for Sir3 binding (C) and Sir1 binding (D) to HMR-SSa by ChIPs. Error bars indicate standard deviations.
Orc1BAH is not the only BAH domain implicated in silencing. In fact, the protein in yeast most similar to Orc1 is the silencing protein Sir3. The strong similarity over the Orc1 and Sir3 proteins' BAH domains raises the possibility that these two domains perform similar functions (3). However, there is little compelling evidence for a direct and specific interaction between Sir1 and the Sir3BAH domain. Sir3BAH does not interact with Sir1OIR because it lacks key surface-exposed residues critical to the Sir1OIR-Orc1BAH complex (13, 31; Z. Hou, unpublished data). However, we reasoned that since Sir125-155 contains a region similar to OIR (13) (Fig. 5A and B), its biologically relevant interaction partner might be Sir3BAH. If this is correct, then Sir125-155-GBD should interact well with GAD-Sir3BAH. However, although a two-hybrid assay indicated that a GAD-Sir3BAH fusion did indeed interact with Sir125-155-GBD, it was only slightly more effective at doing so than a GAD-Orc1BAH fusion (Fig. 6A). A number of GBD and GAD fusion combinations were examined, including full-length GAD-Sir3 and an N-terminal region of Sir1 that included both N1 and N2, with the same basic result: both the Orc1BAH and Sir3BAH domains interacted weakly with the N-terminal region of Sir1 (L. Mendoza, unpublished data). This weak and relatively nonselective interaction contrasted sharply with that of the Sir1OIR and Orc1BAH; GBD-Sir1473-678 interacted robustly with GAD-Orc1BAH but not detectably with GAD-Sir3BAH (Fig. 6A). Thus, the Sir1 N-terminal region interacted weakly with both the Orc1 and Sir3 BAH domains but showed little selectivity for one domain over the other.
Although the two-hybrid data supporting a Sir1 N-terminal-Sir3BAH interaction were not compelling, they did not rule out the possibility that the Sir3BAH domain mediated the roles of N1 and N2 in stabilizing Sir1 binding to HMRa in vivo. Perhaps a weak interaction between Sir3BAH and the Sir1 N terminus was sufficient to help Sir1 bind HMRa; Sir1's ability to bind HMR-SSa does require the other SIR genes, including SIR3 (Fig. 2E). Even at natural HMR and HML loci, Sir1 binding is reduced in sir3Δ mutant cells (47), albeit not abolished as it is at HMR-SSa. In addition, some genetic data are consistent with an interaction between the Sir1 N terminus and Sir3BAH (13). If a Sir3BAH-Sir1-N-terminus interaction mediated the roles that N1 and N2 played in Sir1 binding to HMR-SSa, then the Sir3BAH domain should be required for wild-type Sir1 to bind HMR-SSa. Therefore, we performed Sir1-directed ChIPs in cells containing a sir3BAHΔ allele substituted for SIR3 at its native locus. The sir3BAHΔ allele contained a deletion of the N-terminal region of Sir3 encoding amino acids 2 to 229.
The sir3BAHΔ allele allowed for expression of sir3BAHΔ protein at levels similar to that of wild-type Sir3 (Fig. 6B). As expected (3), sir3BAHΔ failed to silence HMR-SSa or telomeres (J. R. Danzer, unpublished data). In addition, sir3BAHΔ failed to bind HMR-SSa as measured by ChIP (Fig. 6C). Nevertheless, and unexpectedly based on these observations, sir3BAHΔ was not equivalent to sir3Δ in terms of Sir1 binding to HMR-SSa. Whereas a sir3Δ virtually abolished Sir1 binding to HMR-SSa, a sir3BAHΔ allowed Sir1 to bind to HMR-SSa at levels similar to wild type (Fig. 6D). At present we can only speculate on why a version of Sir3 that was incapable of silencing or binding HMR-SSa could restore Sir1 binding to HMR-SSa. Perhaps the BAH domain-independent chromatin binding capability of Sir3 is relevant (1, 7). Nevertheless, the most relevant point was that the Sir3BAH domain was not the determinant for the roles of N1 and N2 in Sir1's binding to HMR-SSa.
Full-length Sir1 is less effective than Sir1OIR in binding Orc1BAH in vitro.
A conundrum created by the two-hybrid data with full-length GBD-Sir1 was that the C-terminal region of Sir1, completely lacking Sir1's first 472 amino acids (and thus lacking N1 and N2), is fully capable of interacting with Orc1BAH (Fig. 5G, GBD-sir1473-678) (4), yet amino acid substitutions in the N1 and N2 regions of Sir1 substantially reduced the two-hybrid interaction between full-length Sir1 fusions and Orc1BAH (Fig. 4B and 5G). These paradoxical data raised the following question: how could amino acid substitutions in N1 or N2, regions far removed from the Sir1OIR prevent an interaction between the Sir1OIR and Orc1BAH domains when the entire Sir1 N terminus is completely unnecessary for this interaction?
One explanation was that the wild-type N-terminal region of Sir1 occludes the Sir1OIR to some degree, reducing its ability to interact with Orc1BAH, and that the OIR-similar region, perhaps via weak interactions with a BAH domain, is needed to relieve this intramolecular inhibition. That is, defects in N1 and possibly N2 inhibit the Sir1-Orc1BAH interaction because they are necessary to reduce the Sir1 N-terminal region's occlusion of the Sir1OIR. This explanation leads to a prediction: full-length Sir1 should interact less well than Sir1OIR with the Orc1BAH domain. This possible explanation was supported by the two-hybrid data shown in Fig. 5G; GBD-sir1473-678 interacted about 10-fold more efficiently than wild-type full-length GBD-Sir1 with GAD-Orc1BAH even though GBD-sir1473-678 produced less protein than GBD-Sir1 (Fig. 5F). To begin to address this issue in a more defined context, we purified recombinant Sir1 from Escherichia coli and determined whether it was capable of binding to Orc1BAH in a glutathione S-transferase (GST) fusion protein affinity experiment (Fig. 7A). Sir1 bound efficiently to GST-Orc1BAH but not GST alone, suggesting that we had purified functional Sir1 (Fig. 7A). Next we compared the abilities of full-length Sir1 and the Sir1OIR to form a stable complex with Orc1BAH using gel filtration experiments (Fig. 7B). In these experiments the same molar concentration of either Sir1 or Sir1OIR (final concentration, 12 μM) was mixed with an excess of Orc1BAH (final concentration, 20 μM) and incubated for 30 min at 4°C. These concentrations were above the Kd determined in Fig. 1 for Sir1OIR-Orc1BAH, and previous experiments indicated that under these conditions, a Sir1OIR-Orc1BAH complex forms efficiently, with virtually all of the Sir1OIR existing in a Sir1OIR-Orc1BAH complex that can be isolated by gel filtration (30). The mixtures were then fractionated over a Superdex200 gel filtration column, and eluted fractions were analyzed by SDS-PAGE (Fig. 7B). Virtually all of the Sir1OIR coeluted with Orc1BAH, indicative of a stable Sir1OIR-Orc1BAH complex, as expected. In contrast, however, only a small fraction of Orc1BAH coeluted with the larger full-length Sir1. These data provided evidence that the Sir1-Orc1BAH complex formed less efficiently and/or was less stable than the Sir1OIR-Orc1BAH complex. Thus, the presence of the Sir1 N-terminal half of Sir1 within full-length Sir1 impeded the Sir1OIR-Orc1BAH interaction in vitro.
The in vitro data in Fig. 7B and the two-hybrid data in Fig. 4 and 5 suggested that the Sir1OIR was less accessible to binding the Orc1BAH domain within the context of full-length Sir1. The Sir1OIR itself is a discrete and stable globular domain resistant to limited proteolysis (4). If full-length Sir1 existed in a structure in which the Sir1OIR was inaccessible to Orc1BAH, then an expectation was that full-length Sir1 should form a large, relatively stable domain that included both the N terminus and the “protected” Sir1OIR. To test this possibility, we performed a limited trypsin digest of full-length purified Sir1 (Fig. 7C). These experiments revealed that full-length Sir1 did indeed contain a large trypsin-resistant fragment of ∼71 kDa (Fig. 7C, band 1). With increasing incubation times, additional kinetically stable protein fragments were generated (bands 2 and 3). We performed N-terminal protein sequencing on these fragments, which revealed that each contained the Sir1OIR and various amounts of additional N-terminal sequence (Fig. 7D). These data provided evidence that the Sir1OIR was contained within a larger stable domain of Sir1 that included the N-terminal region of the protein.
DISCUSSION
Phylogenetic conservation and homology modeling provides evidence that the Sir1 N terminus is functionally important.
The Sir1 N terminus and the Sir1OIR, a known Orc1BAH interaction domain required for silencing (30, 32), share significant sequence similarity, raising the possibility that the Sir1 N terminus also interacts with a BAH domain-containing protein partner, such as Orc1 or Sir3 (13). In addition, sequence conservation between SIR1 genes from two distantly related Saccharomyces species, S. cerevisiae and S. castellii, is consistent with the Sir1 N terminus having functional relevance. In particular, alignments between two relatively diverged Sir1 proteins in these species revealed that they shared four short stretches of amino acid identity (defined as invariant contiguous stretches of five or more amino acids). Two stretches lie so close together that they were considered a single conserved region and were contained within the functionally important SRD module. The SRD module within the Sir1OIR makes direct contacts with the Orc1BAH (30, 32). Thus, a key functional module within Sir1 is one of the most conserved regions shared between these diverged Sir1 proteins.
Two additional regions of amino acid identity, N1 and N2, were contained within the Sir1 N terminus. In a structural model of the Sir1 N terminus guided by the sequence similarity that the Sir1 N terminus shares with Sir1OIR, one of these regions, N1, forms a structured module similar to the SRD module. The sequence and predicted structural conservation supported the idea that the Sir1 N terminus was functionally important and raised the possibility that its role was mediated, at least in part, through interactions with a BAH domain-containing partner protein(s).
N1 and N2 are required for silencing.
Two different alleles of SIR1 that contained mutations within the N1 and N2 coding regions (sir1n1 and sir1n2) produced mutant Sir1 proteins that failed to silence or bind HMR-SSa. Thus, N1 and N2 were required for Sir1 to function in silencing. To strengthen this conclusion, multiple independent sir1 alleles that targeted N1 were generated using the structural model of the Sir1 N terminus as a guide. Each of these new sir1 alleles produced the same mutant phenotypes as sir1n1 and sir1n2. In particular, when expressed at chromosomal levels, each allele produced a mutant Sir1 protein that failed to either silence or bind HMR-SSa. Thus, five independent alleles affecting the Sir1 N terminus behaved like the previously characterized sir1srd alleles when expressed at chromosomal levels (4, 21, 50). However, in contrast to sir1srd alleles, the alleles affecting the Sir1 N terminus (sir1n) restored full silencing when overexpressed. Thus, the N1 and N2 regions were not absolutely essential for Sir1 to silence HMRa.
The Sir1 N terminus has both positive and negative roles.
The genetic analyses of mutant sir1 alleles that affected the N1 and N2 regions, as well as analysis of a sir1NΔ allele, established that the Sir1 N terminus had a positive role in silencing. However, the two-hybrid experiments provided evidence that the Sir1 N terminus might also possess a negative role in modulating the formation of a Sir1OIR-Orc1BAH complex necessary for silencing. Specifically each of the five alleles affecting the Sir1 N terminus produced mutant Sir1 proteins that showed a reduced interaction with Orc1BAH in a two-hybrid assay. These data were unexpected because these proteins contain a fully functional Sir1OIR that is sufficient to interact with the Orc1BAH. Thus, the Sir1 N terminus was dispensable for a Sir1-Orc1BAH two-hybrid interaction, and yet specific amino acid substitutions within the N1 or N2 regions could substantially reduce this interaction. Importantly, these exact same mutant proteins were capable of silencing HMR-SSa when overexpressed through two different established Sir1-silencing mechanisms (the “natural” mechanism and the Gal4-tethered mechanism), suggesting that these alleles did not produce mutant Sir1 proteins with extremely gross structural defects. (It is probable that the two-hybrid assay requires a more efficient Sir1-Orc1BAH interaction to produce a positive signal than the silencing assay; silencing could be achieved by overexpressing the sir1n1 and sir1n2 mutants even though wild-type levels of Sir1 binding to the silencer and a Sir1-Orc1BAH two-hybrid interaction were not produced.)
In addition, two other pieces of data supported the notion that the Sir1 N terminus had an inhibitory role in Sir1 function. First, a sir1NΔ allele produced ∼10-fold-greater silencing than the sir1n1 or sir1n2 allele as measured by quantitative mating. Second, full-length Sir1 was less efficient at forming a stable complex with Orc1BAH in vitro than Sir1OIR, consistent with the Sir1 N terminus inhibiting access of the Sir1OIR to Orc1BAH.
A speculative explanation consistent with the paradoxical two-hybrid data and other data presented in this study is that Sir1 can exist in two different conformations, one of which occludes access of the Sir1OIR for binding by the Orc1BAH domain (Fig. 7E). In this model, a weak interaction between the Sir1 N terminus and a BAH domain enhances a conformation of Sir1 that presents the Sir1OIR for a robust Sir1OIR-Orc1BAH interaction. The relevant BAH partner for the Sir1 N terminus is unknown, but in the context of the two-hybrid assay with full-length GBD-Sir1 as bait, the Orc1BAH domain played this role. Thus, in the two-hybrid assay with full-length Sir1, the Orc1BAH domain had two roles: (i) it interacted weakly with the Sir1 N terminus to promote a conformation of Sir1 in which the Sir1OIR was exposed, and (ii) it bound to the Sir1OIR with a high affinity. (The Orc1BAH molecule that interacts with a given Sir1 to trigger a conformational change need not be [and likely is not] the same Orc1BAH molecule that then binds that Sir1.) In the context of native silencing, the BAH partner necessary for the first step may not necessarily be Orc1BAH. Regardless, if this model was correct and the masking of the Sir1OIR was a natural by-product of native Sir1 structure, then the Sir1OIR should interact more effectively with Orc1BAH than full-length Sir1 in vitro because a fraction of Sir1 molecules in a given population should exist in a conformation in which the Sir1OIR was masked. This result was observed in our in vitro analysis.
We have also considered an alternative, less “complicated” model where the sir1n mutations that we generated produced an altered protein structure that caused the N terminus to “collapse” onto Sir1OIR and occlude it from interacting with Orc1BAH. However, in this scenario one would predict that wild-type Sir1 and the Sir1OIR would interact equally well with Orc1BAH in vitro, which was not what we observed. In addition, we would expect that such a protein structural defect would have produced less functional versions of Sir1 rather than the hypomorphic versions observed in this study. Thus, although we cannot rule out this alternative model and many additional experiments are necessary to challenge the model in Fig. 7E, we currently favor it as the simplest explanation that incorporates all of the data.
It is important to note that while this model addresses the potential negative and positive roles of the Sir1 N terminus in controlling access of Orc1BAH to the Sir1OIR, it does not directly address the positive role of the Sir1 N terminus in stabilizing Sir1's association with HMRa once Sir1 has bound ORC (i.e., once a stable Sir1OIR-Orc1BAH complex has formed). Regardless of how the Sir1 N terminus affects the Sir1OIR-Orc1BAH interaction, which is the concern of the model presented in Fig. 7E, it is probable that it also interacts with other proteins at HMRa to stabilize Sir1 binding to this locus.
Confining Sir1 to silencers.
The role of inhibitory domains in modulating protein function is well documented in biology. For example, protein kinase A activity is controlled by regulatory domains (53), as is the ability of steroid hormone receptors to bind DNA (5). In these famous examples, small ligands, produced in response to physiologically relevant stimuli, elicit the protein conformational changes necessary to change protein activity. At this stage of our understanding of silencing, it is less clear how a conformational change in Sir1 that promotes its interaction with ORC might be relevant, but it is worth noting that inhibitory domains within other Sir proteins have been documented. For example a region(s) within the N-terminal two-thirds of Sir4 inhibits interactions between Sir4 and Sir3 (40). In addition, in vitro, the conformation of a Sir2-Sir4 complex is changed by O-acetyl ADP-ribose (37). Thus, modulating protein conformations may be a general feature affecting Sir proteins and other proteins involved in complex higher-order chromatin structures.
In terms of Sir1-ORC interactions, we speculate that conformational changes in Sir1 might contribute to the selectivity that Sir1 shows for silencer-bound ORCs over origin-bound ORCs in vivo (21). Perhaps a productive interaction between the Sir1 N terminus and a BAH domain would be more likely to occur near HMRa because this locus is naturally associated with a greater number of BAH domain containing proteins compared to other regions of the genome that bind ORC (i.e., origins). For example, Sir3 is associated with HMRa and HMLα to some degree even in the absence of SIR1, and even though Sir3BAH was not necessary for Sir1 binding to HMR-SSa, this could be because its role overlapped with that of Orc1BAH or even another BAH partner protein (47). In addition HMRa is one of the unusual regions of the genome in terms of ORC binding since it is highly enriched for ORCs compared to other regions, based on genome-wide binding studies (56). Therefore, perhaps when Sir1 is near the HM loci, the probability that its Sir1OIR would be made accessible for binding to Orc1BAH through a BAH-induced conformational change would be higher than when it is near an origin. Alternatively, the conformational change that we propose could occur much earlier in the formation of silencer-binding subcomplexes that include Sir1 and ORC and other Sirs that together show high affinity for the DNA sequence composition of silencers. In either scenario, selectivity is achieved by regulating the formation and/or nature of a Sir1-ORC complex.
Does the N terminus interact with a BAH domain?
Regardless of how the Sir1 N terminus works within Sir1, our thinking has been influenced by the idea that its function(s) involve an interaction(s) with a BAH domain-containing protein. First, there is the strong similarity between the Sir1 N-terminal region containing N1 and the Sir1OIR, a known BAH domain interaction partner. Second, two-hybrid data provide molecular evidence that the Sir1 N terminus interacts weakly with the Sir3 and Orc1 BAH domains and other BAH domains (L. Mendoza, unpublished data). However, the interactions that we have observed to date are weak and show little selectivity for a particular BAH domain partner. Perhaps a weak interaction that is productive (i.e., promotes a conformational change in Sir1 to expose the Sir1OIR) in only a fraction of all protein-protein contacts may be exactly what is needed to achieve a selective interaction between Sir1 and a silencer-bound ORC. Alternatively, a BAH domain might be only one of two or possibly more protein domains that come together to form a Sir1 N terminus interaction surface. For example, Sir4 is a potential candidate protein for interactions with the Sir1 N terminus. In particular, residues in the Sir1OIR were needed for a Sir1-Sir4 interaction. Thus, some of the similarity between the Sir1 N terminus's OIR-similar region and the Sir1OIR may be relevant to Sir1-Sir4 interactions. Future experiments will address precisely how Sir1 structure influences its specialized functions in targeting and stabilizing silent chromatin formation to discrete genomic regions through interactions with the multifunctional ORC.
Acknowledgments
We thank Erika Shor for comments on the manuscript and all other members of the lab for useful discussions about these experiments. We thank Patricia J. Kiley and James L. Keck for discussions about portions of this work. We thank John M. Denu and members of his lab for help with isothermal titration calorimetry experiments.
This work was supported by funding from the National Institutes of Health in the forms of an NRSA postdoctoral fellowship (GM072392 to J.R.D.) and an RO1 grant (GM056890 to C.A.F.). C.A.F. is also grateful for supplemental support from the UW Madison Graduate School and VILAS Associate and Life Cycle Awards.
Footnotes
Published ahead of print on 24 November 2008.
REFERENCES
- 1.Altaf, M., R. T. Utley, N. Lacoste, S. Tan, S. D. Briggs, and J. Cote. 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 281002-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16659-672. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P., J. Mitchell, J. Leber, R. Kobayashi, and B. Stillman. 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83563-568. [DOI] [PubMed] [Google Scholar]
- 4.Bose, M. E., K. H. McConnell, K. A. Gardner-Aukema, U. Muller, M. Weinreich, J. L. Keck, and C. A. Fox. 2004. The origin recognition complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Mol. Cell. Biol. 24774-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brent, G. A., D. D. Moore, and P. R. Larsen. 1991. Thyroid hormone regulation of gene expression. Annu. Rev. Physiol. 5317-35. [DOI] [PubMed] [Google Scholar]
- 6.Callebaut, I., J. C. Courvalin, and J. P. Mornon. 1999. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446189-193. [DOI] [PubMed] [Google Scholar]
- 7.Carmen, A. A., L. Milne, and M. Grunstein. 2002. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J. Biol. Chem. 2774778-4781. [DOI] [PubMed] [Google Scholar]
- 8.Casey, L., E. E. Patterson, U. Muller, and C. A. Fox. 2008. Conversion of a replication origin to a silencer through a pathway shared by a forkhead transcription factor and an S phase cyclin. Mol. Biol. Cell 19608-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien, C. T., S. Buck, R. Sternglanz, and D. Shore. 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75531-541. [DOI] [PubMed] [Google Scholar]
- 10.Chivian, D., and D. Baker. 2006. Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic Acids Res. 34e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chivian, D., D. E. Kim, L. Malmstrom, P. Bradley, T. Robertson, P. Murphy, C. E. Strauss, R. Bonneau, C. A. Rohl, and D. Baker. 2003. Automated prediction of CASP-5 structures using the Robetta server. Proteins 53(Suppl. 6)524-533. [DOI] [PubMed] [Google Scholar]
- 12.Chivian, D., D. E. Kim, L. Malmstrom, J. Schonbrun, C. A. Rohl, and D. Baker. 2005. Prediction of CASP6 structures using automated Robetta protocols. Proteins 61(Suppl. 7)157-166. [DOI] [PubMed] [Google Scholar]
- 13.Connelly, J. J., P. Yuan, H. C. Hsu, Z. Li, R. M. Xu, and R. Sternglanz. 2006. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol. Cell. Biol. 263256-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cubizolles, F., F. Martino, S. Perrod, and S. M. Gasser. 2006. A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol. Cell 21825-836. [DOI] [PubMed] [Google Scholar]
- 15.Delano, W. L. 2002. The PyMol molecular graphics system. DeLano Scientific, San Carlos, CA.
- 16.Denu, J. M. 2003. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem. Sci. 2841-48. [DOI] [PubMed] [Google Scholar]
- 17.Dhillon, N., and R. T. Kamakaka. 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell 6769-780. [DOI] [PubMed] [Google Scholar]
- 18.Fox, C. A., A. E. Ehrenhofer-Murray, S. Loo, and J. Rine. 1997. The Origin recognition complex, SIR1, and the S phase requirement for silencing. Science 2761547-1551. [DOI] [PubMed] [Google Scholar]
- 19.Fox, C. A., S. Loo, A. Dillin, and J. Rine. 1995. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev. 9911-924. [DOI] [PubMed] [Google Scholar]
- 20.Fox, C. A., and K. H. McConnell. 2005. Toward biochemical understanding of a transcriptionally silenced chromosomal domain in Saccharomyces cerevisiae. J. Biol. Chem. 2808629-8632. [DOI] [PubMed] [Google Scholar]
- 21.Gardner, K. A., and C. A. Fox. 2001. The Sir1 protein's association with a silenced chromosome domain. Genes Dev. 15147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner, K. A., J. Rine, and C. A. Fox. 1999. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics 15131-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasser, S. M., and M. M. Cockell. 2001. The molecular biology of the SIR proteins. Gene 2791-16. [DOI] [PubMed] [Google Scholar]
- 24.Gavin, K. A., M. Hidaka, and B. Stillman. 1995. Conserved initiator proteins in eukaryotes. Science 2701667-1671. [DOI] [PubMed] [Google Scholar]
- 25.Georgel, P. T., M. A. Palacios DeBeer, G. Pietz, C. A. Fox, and J. C. Hansen. 2001. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl. Acad. Sci. USA 988584-8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodwin, G. H., and R. H. Nicolas. 2001. The BAH domain, polybromo and the RSC chromatin remodelling complex. Gene 2681-7. [DOI] [PubMed] [Google Scholar]
- 27.Grunstein, M. 1997. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 9383-387. [DOI] [PubMed] [Google Scholar]
- 28.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA.
- 29.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Hou, Z., D. A. Bernstein, C. A. Fox, and J. L. Keck. 2005. Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proc. Natl. Acad. Sci. USA 1028489-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou, Z., J. R. Danzer, C. A. Fox, and J. L. Keck. 2006. Structure of the Sir3 protein bromo adjacent homology (BAH) domain from S. cerevisiae at 1.95 A resolution. Protein Sci. 151182-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu, H. C., B. Stillman, and R. M. Xu. 2005. Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing. Proc. Natl. Acad. Sci. USA 1028519-8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1441425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, D. E., D. Chivian, and D. Baker. 2004. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 32W526-W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitazono, A. A., B. T. Tobe, H. Kalton, N. Diamant, and S. J. Kron. 2002. Marker-fusion PCR for one-step mutagenesis of essential genes in yeast. Yeast 19141-149. [DOI] [PubMed] [Google Scholar]
- 36.Leatherwood, J., and A. Vas. 2003. Connecting ORC and heterochromatin: why? Cell Cycle 2573-575. [PubMed] [Google Scholar]
- 37.Liou, G. G., J. C. Tanny, R. G. Kruger, T. Walz, and D. Moazed. 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121515-527. [DOI] [PubMed] [Google Scholar]
- 38.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 39.McNally, F. J., and J. Rine. 1991. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 115648-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moazed, D., A. Kistler, A. Axelrod, J. Rine, and A. D. Johnson. 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 942186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moazed, D., A. D. Rudner, J. Huang, G. J. Hoppe, and J. C. Tanny. 2004. A model for step-wise assembly of heterochromatin in yeast. Novartis Found. Symp. 25948-56. (Discussion, 259:56-62, 163-169.) [PubMed] [Google Scholar]
- 42.Onishi, M., G. G. Liou, J. R. Buchberger, T. Walz, and D. Moazed. 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 281015-1028. [DOI] [PubMed] [Google Scholar]
- 43.Palacios DeBeer, M. A., and C. A. Fox. 1999. A role for a replicator dominance mechanism in silencing. EMBO J. 183808-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohl, C. A., C. E. Strauss, D. Chivian, and D. Baker. 2004. Modeling structurally variable regions in homologous proteins with Rosetta. Proteins 55656-677. [DOI] [PubMed] [Google Scholar]
- 45.Rudner, A. D., B. E. Hall, T. Ellenberger, and D. Moazed. 2005. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 254514-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72481-516. [DOI] [PubMed] [Google Scholar]
- 47.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 132207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Sasaki, T., and D. M. Gilbert. 2007. The many faces of the origin recognition complex. Curr. Opin. Cell Biol. 19337-343. [DOI] [PubMed] [Google Scholar]
- 50.Sharp, J. A., D. C. Krawitz, K. A. Gardner, C. A. Fox, and P. D. Kaufman. 2003. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes Dev. 172356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stone, E. M., M. J. Swanson, A. M. Romeo, J. B. Hicks, and R. Sternglanz. 1991. The SIR1 gene of Saccharomyces cerevisiae and its role as an extragenic suppressor of several mating-defective mutants. Mol. Cell. Biol. 112253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor, S. S., J. A. Buechler, and W. Yonemoto. 1990. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 59971-1005. [DOI] [PubMed] [Google Scholar]
- 54.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56619-630. [DOI] [PubMed] [Google Scholar]
- 55.Triolo, T., and R. Sternglanz. 1996. Role of interactions between the origin recognition complex and Sir1 in transcriptional silencing. Nature 381251-253. [DOI] [PubMed] [Google Scholar]
- 56.Xu, W., J. G. Aparicio, O. M. Aparicio, and S. Tavare. 2006. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, Z., M. K. Hayashi, O. Merkel, B. Stillman, and R. M. Xu. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 214600-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]