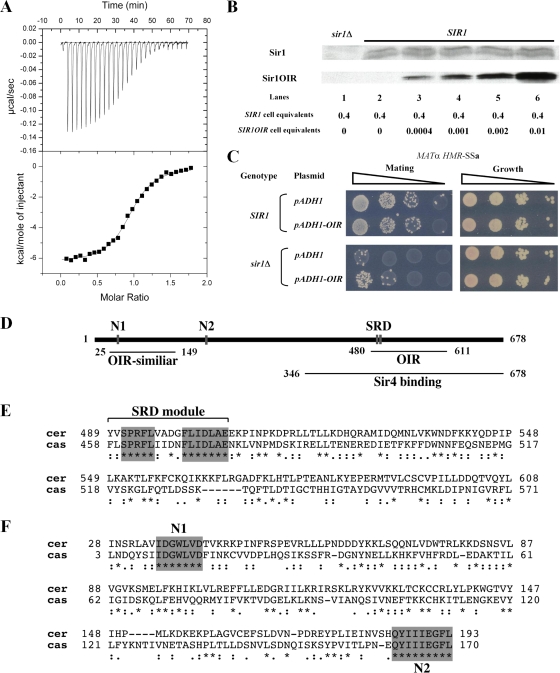
FIG. 1.
Sir1OIR binds Orc1BAH efficiently in vitro but does not compete with wild-type Sir1 for silencing when overexpressed. (A) Binding between Sir1OIR and Orc1BAH was measured by isothermal titration calorimetry. Sir1OIR and Orc1BAH were purified (30) and dialyzed extensively in 25 mM phosphate (pH 7.2)-200 mM NaCl. The protein domains were used at 145 μM for Orc1BAH and 6.5 μM for Sir1OIR. The Kd for the Sir1OIR-Orc1BAH interaction was calculated as 0.20 ± 0.03 μM. (B) Cell extracts from MATα SIR1 (CFY345) yeast were examined for Sir1 and Sir1OIR by protein immunoblotting with an anti-Sir1 antibody. One strain contained a vector carrying the ADH1 promoter (pCF1883); its only source of Sir1 was from the SIR1 locus (SIR1). The second strain was identical except that it contained a plasmid in which the ADH1 promoter drove expression of SIR1OIR fused to an NLS (SIR1OIR) (pCF1881). The extracts were mixed prior to SDS-PAGE at the indicated cell equivalent amounts, expressed in A600 units. The rabbit polyclonal anti-Sir1 used in this experiment was raised against Sir1OIR purified from E. coli. The Sir1OIR-containing-extract was diluted to 0.001 A600 unit to achieve signals comparable to that for 0.4 A600 unit of Sir1 extract (lane 4). The level of native Sir1 in each lane served as internal loading control; in addition, the filter was also stained with Ponceau S prior to immunoblotting to determine levels of protein loading and transfer that would allow interpretation. The Epi Chemi II Dark Room system and LabWorks Analysis software (UVP Laboratory Products) were used for quantification. (C) pADH1 (pCF1883)- or pADH1-NLS-SIR1OIR (pADH1-OIR; pCF1881)-containing plasmids were transformed into MATα HMR-SSa cells that were either SIR1 (CFY345) or sir1Δ (CFY762). Silencing of HMR-SSa was measured in semiquantitative mating assays in which 10-fold serial dilutions of cells being tested were mixed with an excess of MATa cells (CFY616) and plated to selective agar medium (Mating) that allowed only diploid cells to grow. The MATα cells used in the mating assay were simultaneously examined for viability on nonselective agar medium (Growth) to ensure that equivalent numbers of cells were used. (D) Established and potential functional regions of Sir1. The 122-amino-acid OIR binds the Orc1BAH domain, and its structure has been solved (30, 32). The SRD region is conserved between the diverged Sir1 proteins from S. castellii and S. cerevisiae and forms the surface on the Sir1OIR that is involved in direct contacts with Orc1BAH (30, 32). The Sir4 binding region is sufficient to produce a weak two-hybrid interaction with Sir4 (4, 55). The N1 and N2 regions are also highly conserved between these S. castellii and S. cerevisiae Sir1 proteins. A protein BLAST search with the Sir1OIR identifies a region within the Sir1 N terminus as 27% identical and 47% similar (OIR-similar). (E) Protein sequence alignment of Sir1 proteins from S. cerevisiae (top sequence) and S. castellii from amino acid 489 to 608 of S. cerevisiae Sir1. Invariant regions within the SRD module are highlighted. (F) Protein sequence alignment of Sir1 proteins from S. cerevisiae (top sequence) and S. castellii from amino acid 28 to 193 of S. cerevisiae Sir1. The two regions of amino acid identity, N1 and N2, are highlighted. The alignment used here was obtained from the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org/) using the Fungal Alignment option under the Comparison Resources toolbar.