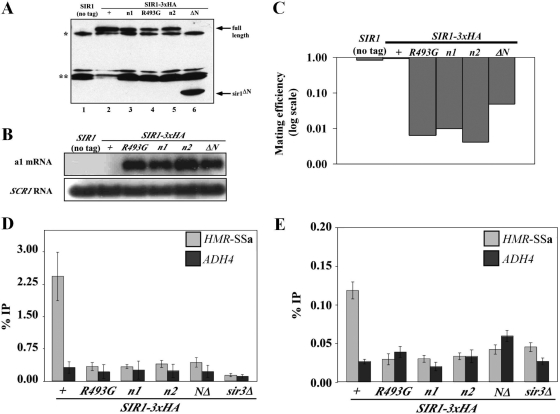
FIG. 2.
N1 and N2 are required for Sir1 to silence and bind HMR. (A) HA-tagged SIR1 alleles were integrated at the SIR1 chromosomal locus, and Sir1-HA3-tagged proteins were detected in protein immunoblots with an anti-HA antibody. * and ** indicate non-Sir1 proteins recognized by anti-HA. Except for the sir1 allele, the yeast cells used in this analysis were isogenic to a MATα HMR-SSa SIR1 strain that was also examined as a control (no tag). (B) RNA blot hybridization of a1 mRNA and SCR1 RNA from yeast cells described in panel A. SCR1 RNA served as a loading control. (C) Quantitative mating assays are performed by measuring the number of cells in a population of viable cells that can mate (and hence silence). The ratio of mating-competent cells to total number of cells is indicated on the y axis (which is shown in log scale) for each strain indicated. (D) Anti-Sir3-directed ChIP experiments. The averages and standard deviations from three independent experiments are shown. ChIPs were performed on a sir3Δ strain (CFY1804) to demonstrate the specificity of the anti-Sir3 antibody. HMR-SSa was detected with HMR-E silencer-specific primers. ADH4 served as a non-Sir1-dependent immunoprecipitation control. Primers used to detect HMR-SSa and ADH4 have been described previously (8). The percentage of HMR-SSa or ADH4 immunoprecipitated out of total starting DNA for each yeast strain is indicated with gray or black bars, respectively. (E) Anti-Sir1 directed ChIP experiments.