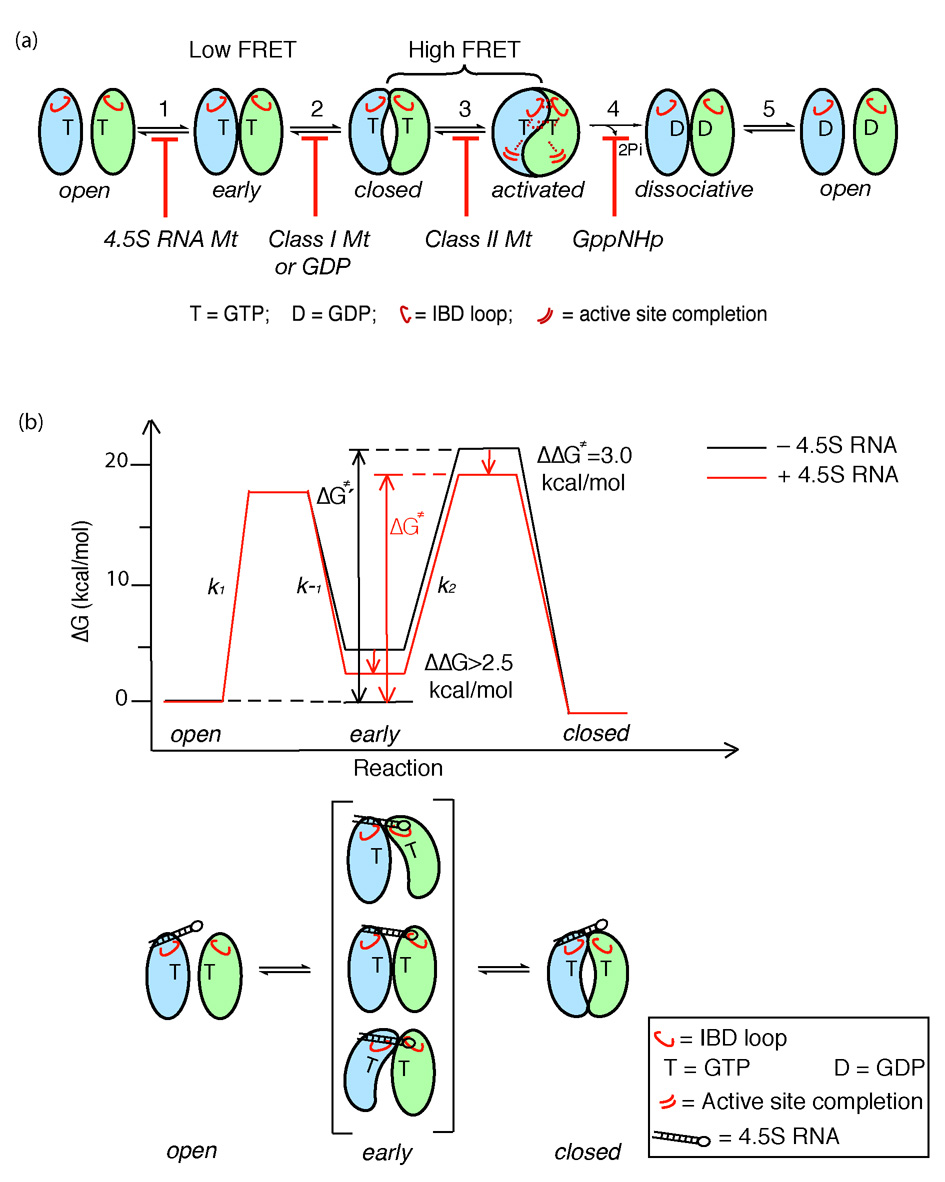
Figure 8. Multiple conformational changes during SRP-SR complex formation and activation.
(a) SRP and SR GTPases form an early GTP-independent intermediate that exhibits a low FRET (step 1). In the presence of GTP, early rearranges to a more stable, closed complex that exhibits a high FRET (step 2). Additional rearrangements in the catalytic loops activate GTP hydrolysis (step 3). GTP hydrolysis drives the dissociation of the SRP•SR complex (steps 4 and 5). Each step can be blocked using specific mutants or nucleotides. 4.5S RNA tetraloop mutants block formation of the early intermediate. Class I mutants of SR 5 or GDP blocks formation of a closed complex. Class II mutants on SRP or SR 5, 6 block the rearrangement that activates GTP hydrolysis. GppNHp blocks the chemical step. (b) top panel: free energy profile for the SRP-SR interaction in the absence (black) and presence (red) of the 4.5S RNA for a standard state of 200 nM that mimics the in vivo protein concentrations in bacteria. Activation energies were calculated from the observed association and dissociation rate constants using ΔG = −RT ln(kh/kBT), where R = 1.987 cal K−1 mol−1, h = 1.58 × 10−37 kcal s−1, kB = 3.3 × 10−27 kcal K−1, and T = 298K. The relative energies of the different complexes were calculated from the observed equilibrium stabilities using ΔG = − RT lnK. The 4.5S RNA stabilizes the early intermediate (in bracket) by > 2.5 kcal mol−1, and the overall activation energy is subsequently lowered by ~3 kcal mol−1. ΔG≠ and ΔG≠′ defines the overall activation energy for forming the GTP-stabilized complex with and without RNA, respectively. The bottom panel depicts a physical picture of how the 4.5S RNA exerts its effect on the SRP-SR interaction as described in the text.