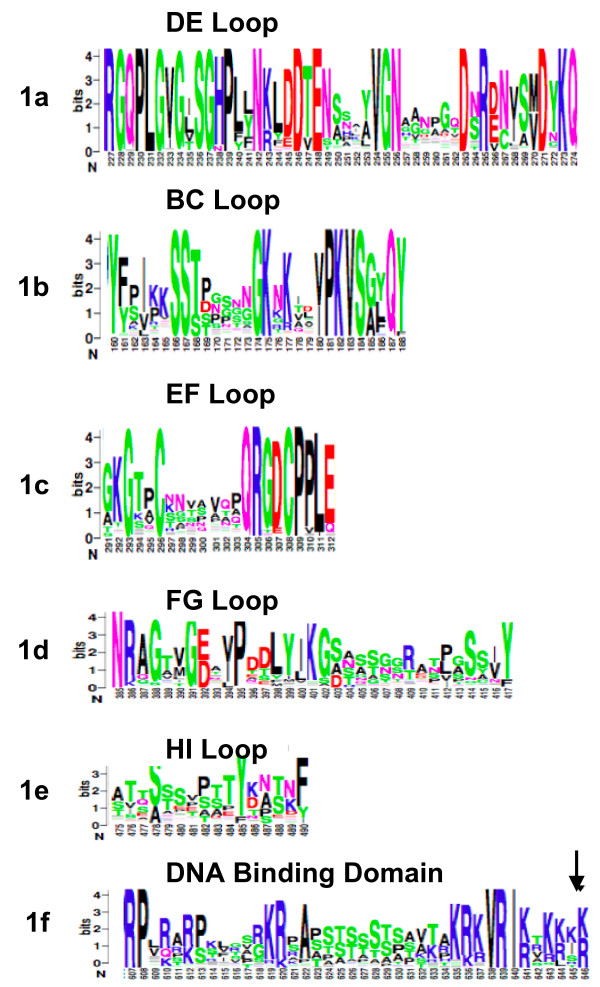
Figure 1.
Analysis of conserved regions within the L1 protein. Seventy-six alpha-HPV L1 sequences were obtained from the NCBI protein database under the Universal Virus Database, as well as six known HPV-16 L1 variant sequences provided by the ICTV. The sequences were aligned using the MUltiple Sequence Comparison by Log-Expectation (MUSCLE). MUSCLE outputs were loaded into CLUSTALX user interface for graphical representation of residue conservation and analysis. Sequence conservation is by the height of residue logos (indicated in bits), as generated by WebLogo 3. The consensus sequences resulting from the alignments of the external loop regions are as follows: DE loop (aa 227–274) (1a), BC (aa 160–188) (1b), EF loop (aa 291–312) (1c), FG loop (aa 385–417) (1d), the HI loop (aa 475–490) (1e) antigenic determinants, and the conserved DNA binding regions (1f). The HPV 16 cysteines at residues 201 and 454, which are involved with disulfide linkages, are conserved across the entire alpha-papillomavirus family.