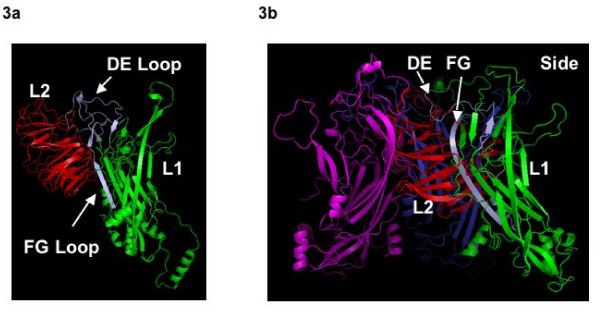
Figure 3.
Predicted 3D model of the L1:L2 interaction. The HPV16 L1 protein structure was obtained from the RCSB Protein Data Bank. The secondary structure of HPV16 L2 protein was predicted with using the 3D-Jigsaw and the Swiss Modelling Server http://swissmodel.expasy.org. The data was then analyzed with the SAM-T09 program (Sequence Alignment and Modeling System, for tertiary structure prediction) which was further refined using AL2TS. The docking position of L2 to L1 was predicted with ClusPro, (Protein-Protein Docking Web Server). The L1 and L2 structures were then visualized using PyMOL, (a molecular visualization program). The predicted L2 structure in its docking position on the L1 monomer (3a). The predicted orientation of the L2 protein within the L1 pentamer structure; two L1 monomers of L1 have been removed to clearly show the alignment of L2 within the structure (3b).