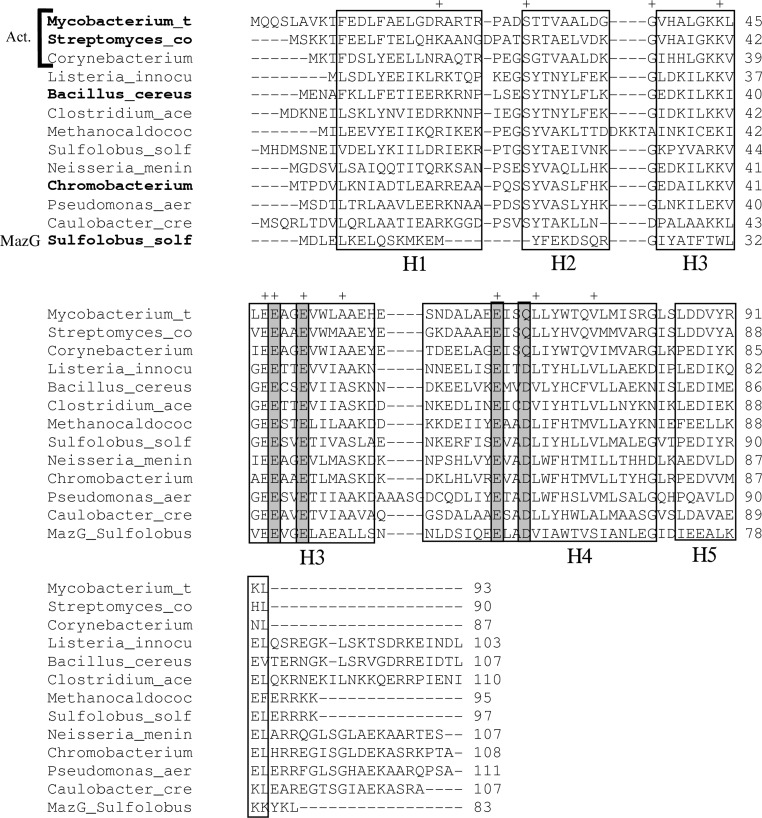
Figure 1.
Multiple alignment of amino-acid sequences of HisE homologs from various bacterial species. The top three rows are from actinobacteria (Act.; Mycobacterium tuberculosis, Streptomyces coelicolor and Corynebacterium glutamicum). The remaining species include Listeria innocua, Bacillus cereus, Clostridium acetobutylicum, Methanococcus jannaschii, Solfolobus solfataricus, Neisseria meningitidis, Chromobacterium violaceum, Pseudomonas aeruginosa and Caulobacter crescentus. A representative MazG sequence from S. solfataricus has also been included for reference (bottom row). The species names in bold indicate proteins for which crystal structures have been solved. The alignment has been adjusted manually to be consistent with known gap placement in these structures. Gray boxes indicate residues that participate in the putative magnesium-binding site. ‘+’ indicates highly conserved residues (identical in at least ten of the 12 HisE sequences).