(b).
Refinement.
| Uncorrected data | Corrected data‡ | |
|---|---|---|
| Molecules in ASU | 2 + 2 monomers | 2 monomers |
| Refined residues | 1540 | 770 |
| Refined waters | 1508 | 754 |
| Rcryst§ | 0.193 | 0.180 |
| Rfree¶ | 0.231 | 0.210 |
| Average B values (Å2) | ||
| Protein | 12.3 | 16.6 |
| Waters | 28.8 | 32.0 |
| Ramachandran plot (%) | ||
| Most favored | 84.2 | 84.3 |
| Additionally allowed | 15.1 | 15.1 |
| Generously allowed | 0.8 | 0.6 |
| Disallowed | 0.0 | 0.0 |
| R.m.s.d. bond lengths (Å) | 0.012 | 0.013 |
| R.m.s.d. bond angles (°) | 1.3 | 1.4 |
R
merge = 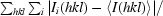
 , where 〈I(hkl)〉 is the average intensity of i symmetry-related observations of reflections with Miller indices hkl.
, where 〈I(hkl)〉 is the average intensity of i symmetry-related observations of reflections with Miller indices hkl.
The refinement statistics for corrected data (PDB code 3beq; Xu et al., manuscript in preparation) are shown here for comparison.
R
cryst = 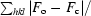
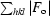 , where F
o and F
c are the observed and calculated structure factors.
, where F
o and F
c are the observed and calculated structure factors.
R free was calculated as for R cryst, but on 5% of data that were excluded prior to refinement.
