Table 2. Comparison of some known protein crystals with lattice-translocation defects.
| Protein† | ϕ29 DNA polymerase | HslV–HslU | S1-RBD-80R | Native 18NA |
|---|---|---|---|---|
| Space group | P21 | H32 | P21 | C2221 |
| Unit-cell parameters (Å, °) | a = 59.9, b = 169.4, c = 68.6, β = 107.6 | a = 181.3, b = 181.3, c = 530.0 | a = 47.5, b = 175.9, c = 67.6, β = 96.6 | a = 117.7, b = 138.5, c = 117.9 |
| Resolution (Å) | 2.8 | 4.2 | 2.3 | 1.65 |
| Average I/σ(I) | 7.6 | 14.7 | 8.8 | 22.2 |
| Completeness (%) | 100 | 100 | 93.8 | 97.3 |
| Rmerge‡ | 0.171 | 0.170 | 0.145 | 0.093 |
| Patterson peak location | (0, 0, 1/2) | (0, 0, 1/3) | (1/3, 0, 0) | (0, 1/2, 1/2) |
| Patterson peak ratio§ (%) | 27 | 26 | 65 | 54 |
| Defect fractions (%) | 83 and 17 | 79 and 21 | 73 and 27 | 77 and 23 |
| Observed ‘streaky’ axis | a* | c* | a* | b* |
Data sources: ϕ29 DNA polymerase, Wang, Kamtekar et al. (2005 ▶); HslV–HslU, Wang, Rho et al. (2005 ▶); S1-RBD-80R, Hwang et al. (2006 ▶).
R
merge = 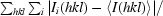
 , where 〈I(hkl)〉 is the average intensity of i symmetry-related observations of reflections with Miller indices hkl.
, where 〈I(hkl)〉 is the average intensity of i symmetry-related observations of reflections with Miller indices hkl.
The Patterson peak ratio is the ratio between the off-origin peak height and the origin peak height.
