Figure 2.
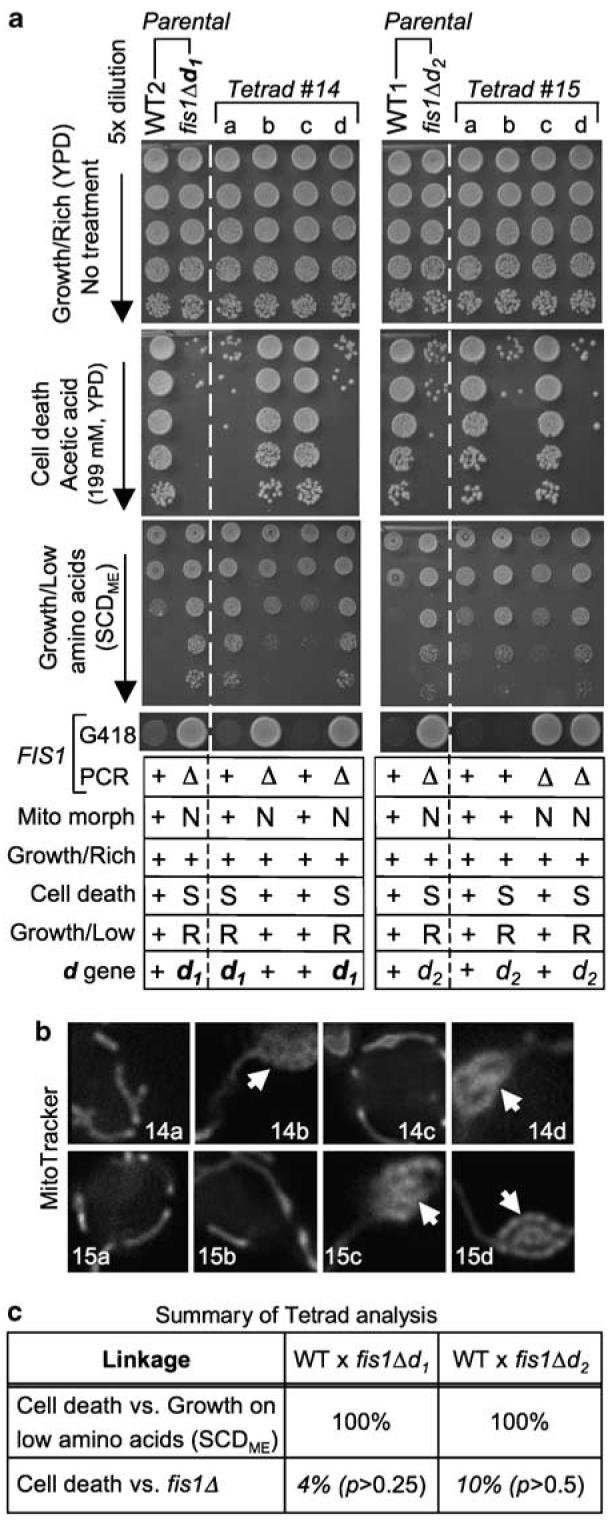
Tetrad analysis reveals a second mutation responsible for cell death and growth control defects. (a) Haploid parental strains and the four spores/strains of a representative tetrad derived from their cross was analyzed for growth on rich medium (YPD, top), tubular ( +/normal) or netted (N) mitochondrial morphology, sensitivity (S) to acetic acid-induced cell death, resistance (R) to growth inhibition due to low amino acids (synthetic complete dextrose medium, SCDME, Methods in Enzymology), and G418 resistance (indicating FIS1 gene replacement). Data are summarized for three or more independent experiments per condition in the table below; + indicates wild-type phenotype. (b) Mitochondrial morphology of each spore in tetrads no. 14 and no. 15 from (a) were visualized by MitoTracker staining.10 Arrows mark netted mitochondrial morphology. (c) Summary of genetic linkage determined by tetrad analyses as illustrated in (a). FIS1 deletion is unlinked to the cell death d gene, as the ratios of PD (parental ditype): NPD (nonparental ditype): TT (tetratype) within tetrads were 1 : 6 : 17 for fis1Δd1 (24 tetrads for cell death and/or overgrowth on SCDME) and 2 : 3 : 15 for fis1Δd2 (20 tetrads), which are close to random segregation (1 : 1 : 4, or 17% for PD (linked) versus 83% for NPD+TT (unlinked)). In contrast, cell death sensitivity was 100% linked to resistance to growth inhibition on low amino acids (SCDME), with ratios of 8 : 0 : 0 (fis1Δd1) and 20 : 0 : 0 (fis1Δd2). χ2 goodness-of-fit test was used to determine whether FIS1-deletion (fis1Δ) segregated independently from mutation d (death) at the expected ratio of 1 : 1 : 4 for PD : NPD : T. As none of the p values rejected the null hypothesis, FIS1-deletion segregated independently from mutation d
