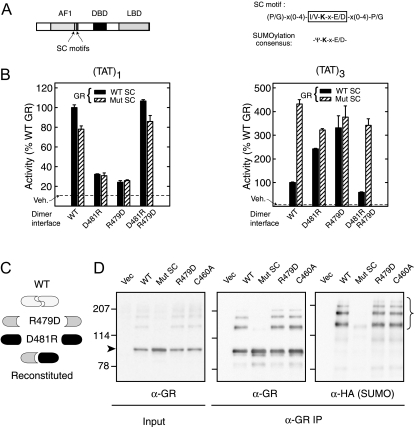
Figure 1.
An Intact GR DBD Dimer Interface Is Required for Synergy Control But Not for SUMOylation
A, Diagram of GR domain structure with SC motifs displayed as vertical lines (left) and SC motif and SUMOylation consensus sequences (right). B, Monkey CV-1 cells were cotransfected with pΔ(TAT)1-Luc (left) or pΔ(TAT)3-Luc (right) reporters together with WT (p6RGR) or synergy control mutant (p6RGR K297R/K313R) GR expression vectors (30 ng) harboring an intact DBD or the salt bridge mutations (R479D or D481R) as indicated. When coexpressed, 15 ng of each mutant was transfected. Data represent averages ± sem of three to four independent transfections performed in triplicate and are expressed as a percentage of the corresponding WT activity (4.1 ± 0.8 and 100.5 ± 19.0 for TAT1 and TAT3, respectively). The activity in vehicle-treated samples was not different among groups and is indicated by the dashed line. The diagram in panel C depicts the effects of the mutant combinations used above with respect to DBD dimerization. D, In vivo SUMOylation. COS-7 cells were cotransfected with expression vectors for HA SUMO1 and for either WT GR (p6RGR), dimerization-deficient GR (R497D), the DNA binding-deficient Zn2+ finger mutant (C460A), or for the synergy control mutant GR (K297R/K313R). Cells were harvested 46 h after transfection including a 10 nm Dex treatment for the last 2 h. Extracts as well as GR immunoprecipitates (BuGR2) were resolved by SDS-PAGE and probed for either GR or HA (SUMO). IP, Immunoprecipitation; Mut, mutant; Vec, vector; Veh, vehicle.